[English] 日本語
 Yorodumi
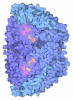
Yorodumi- PDB-7b2c: Crystal structure of the ethyl-coenzyme M reductase from Candidat... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7b2c | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the ethyl-coenzyme M reductase from Candidatus Ethanoperedens thermophilum gassed with xenon | |||||||||
 Components Components | (Ethyl-Coenzyme M reductase ...) x 3 | |||||||||
 Keywords Keywords | TRANSFERASE / Ethyl-CoM reductase / Methyl-CoM reductase / xenon-derivatization / ethane-oxidizers / F430-cofactor / post-translational modification / gas channel / coenzyme M / coenzyme B / true atomic resolution / thermophile / archaea. | |||||||||
| Function / homology | 1-THIOETHANESULFONIC ACID / : / Coenzyme B / Dimethylated-F430 cofactor / Chem-UWT / XENON Function and homology information Function and homology information | |||||||||
| Biological species |  Candidatus Ethanoperedens thermophilum (archaea) Candidatus Ethanoperedens thermophilum (archaea) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.8 Å MOLECULAR REPLACEMENT / Resolution: 1.8 Å | |||||||||
 Authors Authors | Wagner, T. / Lemaire, O.N. / Engilberge, S. | |||||||||
| Funding support |  Germany, 2items Germany, 2items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Crystal structure of a key enzyme for anaerobic ethane activation. Authors: Hahn, C.J. / Lemaire, O.N. / Kahnt, J. / Engilberge, S. / Wegener, G. / Wagner, T. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7b2c.cif.gz 7b2c.cif.gz | 1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7b2c.ent.gz pdb7b2c.ent.gz | 867.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7b2c.json.gz 7b2c.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b2/7b2c https://data.pdbj.org/pub/pdb/validation_reports/b2/7b2c ftp://data.pdbj.org/pub/pdb/validation_reports/b2/7b2c ftp://data.pdbj.org/pub/pdb/validation_reports/b2/7b2c | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7b1sSC  7b2hC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Ethyl-Coenzyme M reductase ... , 3 types, 6 molecules ADBECF
| #1: Protein | Mass: 66344.344 Da / Num. of mol.: 2 / Mutation: wild-type / Source method: isolated from a natural source Details: Seven post-translational modifications exist in the subunit: N1-methylhistidine291 5(S)-methylarginine305 S-methylcysteine354 3-methylisoleucine377 2(S)-methylglutamine445 Thioglycine490 N2-methylhistidine491 Source: (natural)  Candidatus Ethanoperedens thermophilum (archaea) Candidatus Ethanoperedens thermophilum (archaea)Cell line: / / Organ: / / Plasmid details: Culture enrichment / Variant: / / Tissue: / / References: coenzyme-B sulfoethylthiotransferase #2: Protein | Mass: 49874.453 Da / Num. of mol.: 2 / Mutation: wild-type / Source method: isolated from a natural source Source: (natural)  Candidatus Ethanoperedens thermophilum (archaea) Candidatus Ethanoperedens thermophilum (archaea)Cell line: / / Organ: / / Plasmid details: Culture enrichment / Variant: / / Tissue: / / References: coenzyme-B sulfoethylthiotransferase #3: Protein | Mass: 30578.789 Da / Num. of mol.: 2 / Mutation: wild-type / Source method: isolated from a natural source Source: (natural)  Candidatus Ethanoperedens thermophilum (archaea) Candidatus Ethanoperedens thermophilum (archaea)Cell line: / / Organ: / / Plasmid details: Culture enrichment / Variant: / / Tissue: / / References: coenzyme-B sulfoethylthiotransferase |
|---|
-Non-polymers , 12 types, 2405 molecules 







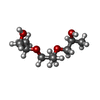














| #4: Chemical | | #5: Chemical | ChemComp-GOL / #6: Chemical | ChemComp-NA / #7: Chemical | #8: Chemical | #9: Chemical | ChemComp-K / #10: Chemical | ChemComp-XE / #11: Chemical | ChemComp-CL / #12: Chemical | #13: Chemical | ChemComp-MG / | #14: Chemical | ChemComp-TRS / | #15: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.28 Å3/Da / Density % sol: 46.06 % / Description: Yellow brick of 200 um long |
|---|---|
| Crystal grow | Temperature: 293.15 K / Method: vapor diffusion, sitting drop / pH: 8.5 Details: Crystals were obtained by initial screening at 20 degree Celsius using the sitting drop method on a 96-Well MRC 2-Drop Crystallization Plates in polystyrene (SWISSCI). The crystallization ...Details: Crystals were obtained by initial screening at 20 degree Celsius using the sitting drop method on a 96-Well MRC 2-Drop Crystallization Plates in polystyrene (SWISSCI). The crystallization reservoir contained 90 ul of mother liquor, crystallization drop contained a mixture of 0.6 ul protein at 16.22 mg.ml-1 and 0.6 ul precipitant. The crystal was obtained by initial screening using the JBScreen Pentaerythritol screen from Jena Bioscience in a Coy tent under an N2/H2 atmosphere (95:5 %). The crystallization reservoir contained 45 % (w/v) Pentaerythritol Propoxylate (5/4 PO/OH), 100 mM Tris pH 8.5 and 400 mM potassium chloride. Crystal was harvested using a MiTeGen MicroRT loop. In order to protect the crystal during xenon pressurisation, a part of the plastic capillary, usually used for room temperature diffraction experiments, was placed around it. The loop was then mounted in an Oxford Xcell xenon chamber. Xenon pressure was gradually increased using a manual pump to reach 25 bars. After 7 min, pressure was slowly released and the crystal immediately plunged in liquid nitrogen. PH range: 7.5 - 8.5 / Temp details: / |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06DA / Wavelength: 1.54981 Å / Beamline: X06DA / Wavelength: 1.54981 Å |
| Detector | Type: DECTRIS PILATUS 2M-F / Detector: PIXEL / Date: Sep 29, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.54981 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→60.87 Å / Num. obs: 232772 / % possible obs: 98.1 % / Redundancy: 6 % / CC1/2: 0.997 / Rmerge(I) obs: 0.105 / Rpim(I) all: 0.046 / Net I/σ(I): 11.7 |
| Reflection shell | Resolution: 1.8→1.84 Å / Redundancy: 4.5 % / Rmerge(I) obs: 0.652 / Mean I/σ(I) obs: 2 / Num. unique obs: 11640 / CC1/2: 0.739 / Rpim(I) all: 0.331 / % possible all: 94.6 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 7B1S Resolution: 1.8→35.9 Å / Cor.coef. Fo:Fc: 0.907 / Cor.coef. Fo:Fc free: 0.888 / SU R Cruickshank DPI: 0.198 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.135 / SU Rfree Blow DPI: 0.118 / SU Rfree Cruickshank DPI: 0.116 Details: LAST REFINEMENT CYCLES WERE PERFORMED WITH HYDROGENS. HYDROGENS WERE OMITTED IN THE FINAL DEPOSITED MODEL.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 91.96 Å2 / Biso mean: 17.66 Å2 / Biso min: 3 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.22 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.8→35.9 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.8→1.82 Å / Rfactor Rfree error: 0 / Total num. of bins used: 51
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj