[English] 日本語
 Yorodumi
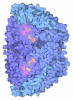
Yorodumi- PDB-5g0r: METHYL-COENZYME M REDUCTASE I FROM METHANOTHERMOBACTER MARBURGENS... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5g0r | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | METHYL-COENZYME M REDUCTASE I FROM METHANOTHERMOBACTER MARBURGENSIS EXPOSED TO 3-NITROOXYPROPANOL | |||||||||
 Components Components | (METHYL-COENZYME M REDUCTASE I SUBUNIT ...) x 3 | |||||||||
 Keywords Keywords | TRANSFERASE / METHYL-COENZYMEM / METHYL-COENZYMEM REDUCTASE EXPOSED TO 3-NITROOXYPROPANOL / POSTTRANSLATIONAL MODIFICATION / BINDING SITES / CATALYSIS / COENZYMES / DISULFIDES / HYDROGEN / LIGANDS / MESNA / METALLOPORPHYRINS / METHANE / METHANOBACTERIUM / NICKEL / OXIDATION-REDUCTION / OXDOREDUCTASES / PHOSPHOTHREONINE / THIOGLYCINE / 3- NITROOXYPROPANOL / INHIBITOR / GREENHOUSE GAS | |||||||||
| Function / homology |  Function and homology information Function and homology informationcoenzyme-B sulfoethylthiotransferase / coenzyme-B sulfoethylthiotransferase activity / methanogenesis / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   METHANOTHERMOBACTER MARBURGENSIS (archaea) METHANOTHERMOBACTER MARBURGENSIS (archaea) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.25 Å MOLECULAR REPLACEMENT / Resolution: 1.25 Å | |||||||||
 Authors Authors | Wagner, T. | |||||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2016 Journal: Proc.Natl.Acad.Sci.USA / Year: 2016Title: Mode of Action Uncovered for the Specific Reduction of Methane Emissions from Ruminants by the Small Molecule 3-Nitrooxypropanol. Authors: Duin, E.C. / Wagner, T. / Shima, S. / Prakash, D. / Cronin, B. / Yanez-Ruiz, D.R. / Duval, S. / Rumbeli, R. / Stemmler, R.T. / Thauer, R.K. / Kindermann, M. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5g0r.cif.gz 5g0r.cif.gz | 1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5g0r.ent.gz pdb5g0r.ent.gz | 846.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5g0r.json.gz 5g0r.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/g0/5g0r https://data.pdbj.org/pub/pdb/validation_reports/g0/5g0r ftp://data.pdbj.org/pub/pdb/validation_reports/g0/5g0r ftp://data.pdbj.org/pub/pdb/validation_reports/g0/5g0r | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5a0yS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||
| Unit cell |
| ||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
-METHYL-COENZYME M REDUCTASE I SUBUNIT ... , 3 types, 6 molecules ADBECF
| #1: Protein | Mass: 60637.648 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: GERMAN COLLECTION OF MICROORGANISMS (DSM) Source: (natural)   METHANOTHERMOBACTER MARBURGENSIS (archaea) METHANOTHERMOBACTER MARBURGENSIS (archaea)Strain: MARBURG References: UniProt: P11558, coenzyme-B sulfoethylthiotransferase #2: Protein | Mass: 47279.676 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: GERMAN COLLECTION OF MICROORGANISMS (DSM) Source: (natural)   METHANOTHERMOBACTER MARBURGENSIS (archaea) METHANOTHERMOBACTER MARBURGENSIS (archaea)Strain: MARBURG References: UniProt: P11560, coenzyme-B sulfoethylthiotransferase #3: Protein | Mass: 28797.234 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: GERMAN COLLECTION OF MICROORGANISMS (DSM) Source: (natural)   METHANOTHERMOBACTER MARBURGENSIS (archaea) METHANOTHERMOBACTER MARBURGENSIS (archaea)Strain: MARBURG References: UniProt: P11562, coenzyme-B sulfoethylthiotransferase |
|---|
-Non-polymers , 7 types, 2638 molecules 











| #4: Chemical | ChemComp-MG / #5: Chemical | #6: Chemical | #7: Chemical | #8: Chemical | ChemComp-K / | #9: Chemical | #10: Water | ChemComp-HOH / | |
|---|
-Details
| Sequence details | RESIDUE 450 IS A DIDEHYDROA |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.18 Å3/Da / Density % sol: 43.6 % / Description: NONE |
|---|---|
| Crystal grow | Temperature: 281 K / Method: vapor diffusion, sitting drop Details: 25-28 % V/V PEG 400, 300 MM MGCL2, 200 MM NACL AND 100 MM HEPES PH 7.5 CRYSTAL GROWTH AT 8 DEGREE CELSIUS UNDER AEROBIC CONDITION |
-Data collection
| Diffraction | Mean temperature: 77 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X10SA / Wavelength: 0.97128 / Beamline: X10SA / Wavelength: 0.97128 |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Mar 9, 2014 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97128 Å / Relative weight: 1 |
| Reflection | Resolution: 1.25→38.79 Å / Num. obs: 633330 / % possible obs: 98.5 % / Observed criterion σ(I): 4 / Redundancy: 4.6 % / Biso Wilson estimate: 9.86 Å2 / Rmerge(I) obs: 0.05 / Net I/σ(I): 14.4 |
| Reflection shell | Resolution: 1.25→1.32 Å / Redundancy: 4.4 % / Rmerge(I) obs: 0.27 / Mean I/σ(I) obs: 4.4 / % possible all: 98.1 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 5A0Y Resolution: 1.25→38.795 Å / SU ML: 0.07 / σ(F): 1.34 / Phase error: 10.8 / Stereochemistry target values: ML Details: CONSIDERING THE RESOLUTION HYDROGEN REFINEMENT MODEL HAS BEEN USED WITH THE RIDING MODE THE INHIBITOR 3-NITROOXYPROPANOL HAS POSSIBLY REDUCED THE NICKEL CENTER AND IS NOW HYDROLYSED IN THE ...Details: CONSIDERING THE RESOLUTION HYDROGEN REFINEMENT MODEL HAS BEEN USED WITH THE RIDING MODE THE INHIBITOR 3-NITROOXYPROPANOL HAS POSSIBLY REDUCED THE NICKEL CENTER AND IS NOW HYDROLYSED IN THE ACTIVE SITE. SINCE THERE IS A LOW PARTIAL OCCUPANCY OF THE BROKEN COMPOUND (NITRITE AND 1-3-PROPANEDIOL) IN THE ACTIVE SITE, WATER MOLECULE HAVE BEEN PLACED INSTEAD.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 15.54 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.25→38.795 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj