[English] 日本語
 Yorodumi
Yorodumi- PDB-5a8k: METHYL-COENZYME M REDUCTASE FROM METHANOTHERMOBACTER WOLFEII AT 1... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5a8k | ||||||
|---|---|---|---|---|---|---|---|
| Title | METHYL-COENZYME M REDUCTASE FROM METHANOTHERMOBACTER WOLFEII AT 1.4 A RESOLUTION | ||||||
 Components Components | (METHYL-COENZYME M ...) x 3 | ||||||
 Keywords Keywords | TRANSFERASE / POST-TRANSLATIONAL MODIFICATION / BINDING SITES / CATALYSIS / COENZYMES / DISULFIDES / HYDROGEN / HYDROGEN BONDING / LIGANDS / MESNA / METALLOPORPHYRINS / METHANE / METHANOBACTERIUM / MODELS / MOLECULAR / NICKEL / OXIDATION-REDUCTION / OXIDOREDUCTASES / PHOSPHOTHREONINE / PROTEIN CONFORMATION / PROTEIN FOLDING / PROTEIN STRUCTURE | ||||||
| Function / homology |  Function and homology information Function and homology informationcoenzyme-B sulfoethylthiotransferase / coenzyme-B sulfoethylthiotransferase activity / methanogenesis / metal ion binding Similarity search - Function | ||||||
| Biological species |   METHANOTHERMOBACTER WOLFEII (archaea) METHANOTHERMOBACTER WOLFEII (archaea) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.41 Å MOLECULAR REPLACEMENT / Resolution: 1.41 Å | ||||||
 Authors Authors | Wagner, T. / Ermler, U. | ||||||
 Citation Citation |  Journal: Angew.Chem.Int.Ed.Engl. / Year: 2016 Journal: Angew.Chem.Int.Ed.Engl. / Year: 2016Title: Didehydroaspartate Modification in Methyl-Coenzyme M Reductase Catalyzing Methane Formation. Authors: Wagner, T. / Kahnt, J. / Ermler, U. / Shima, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5a8k.cif.gz 5a8k.cif.gz | 1014.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5a8k.ent.gz pdb5a8k.ent.gz | 826.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5a8k.json.gz 5a8k.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  5a8k_validation.pdf.gz 5a8k_validation.pdf.gz | 1.6 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  5a8k_full_validation.pdf.gz 5a8k_full_validation.pdf.gz | 1.6 MB | Display | |
| Data in XML |  5a8k_validation.xml.gz 5a8k_validation.xml.gz | 110 KB | Display | |
| Data in CIF |  5a8k_validation.cif.gz 5a8k_validation.cif.gz | 167.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/a8/5a8k https://data.pdbj.org/pub/pdb/validation_reports/a8/5a8k ftp://data.pdbj.org/pub/pdb/validation_reports/a8/5a8k ftp://data.pdbj.org/pub/pdb/validation_reports/a8/5a8k | HTTPS FTP |
-Related structure data
| Related structure data |  5a0yC  5a8rC  5a8wC  3potS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||
| Unit cell |
| ||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
-METHYL-COENZYME M ... , 3 types, 6 molecules ADBECF
| #1: Protein | Mass: 60666.656 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: GERMAN COLLECTION OF MICROORGANISMS (DSM) / Source: (natural)   METHANOTHERMOBACTER WOLFEII (archaea) METHANOTHERMOBACTER WOLFEII (archaea)References: UniProt: H7CHY1*PLUS, coenzyme-B sulfoethylthiotransferase #2: Protein | Mass: 47324.777 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: GERMAN COLLECTION OF MICROORGANISMS (DSM) / Source: (natural)   METHANOTHERMOBACTER WOLFEII (archaea) METHANOTHERMOBACTER WOLFEII (archaea)References: UniProt: A0A1C7D1E2*PLUS, coenzyme-B sulfoethylthiotransferase #3: Protein | Mass: 28727.098 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: GERMAN COLLECTION OF MICROORGANISMS (DSM) / Source: (natural)   METHANOTHERMOBACTER WOLFEII (archaea) METHANOTHERMOBACTER WOLFEII (archaea)References: UniProt: A0A1C7D1E3*PLUS, coenzyme-B sulfoethylthiotransferase |
|---|
-Non-polymers , 8 types, 2512 molecules 



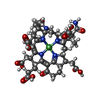
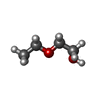









| #4: Chemical | | #5: Chemical | #6: Chemical | ChemComp-K / #7: Chemical | ChemComp-CA / #8: Chemical | #9: Chemical | ChemComp-ETX / #10: Chemical | ChemComp-MG / | #11: Water | ChemComp-HOH / | |
|---|
-Details
| Sequence details | METHANOTHERMOBACTER WOLFEII GENOME SEQUENCING IS ACTUALLY IN PROGRESS AND WILL BE RELEASED SOON. ...METHANOTHE |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.17 Å3/Da / Density % sol: 43.35 % / Description: NONE |
|---|---|
| Crystal grow | Temperature: 281 K / Method: vapor diffusion, sitting drop / pH: 7 Details: 37% (V/V) 2-ETHOXYETHANOL, 50 MM CALCIUM ACETATE, AND 100 MM HEPES PH 7.0 AT 8 DEGREE CELSIUS |
-Data collection
| Diffraction | Mean temperature: 77 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06SA / Wavelength: 0.97798 / Beamline: X06SA / Wavelength: 0.97798 |
| Detector | Type: DECTRIS PILATUS / Detector: PIXEL / Date: Jul 26, 2014 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97798 Å / Relative weight: 1 |
| Reflection | Resolution: 1.41→48.47 Å / Num. obs: 453580 / % possible obs: 99.9 % / Observed criterion σ(I): 2.8 / Redundancy: 8 % / Biso Wilson estimate: 10.09 Å2 / Rmerge(I) obs: 0.12 / Net I/σ(I): 10.7 |
| Reflection shell | Resolution: 1.41→1.49 Å / Redundancy: 7.9 % / Rmerge(I) obs: 0.83 / Mean I/σ(I) obs: 2.8 / % possible all: 99.9 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 3POT Resolution: 1.41→48.467 Å / SU ML: 0.11 / σ(F): 1.34 / Phase error: 12.31 / Stereochemistry target values: ML Details: CONSIDERING THE RESOLUTION HYDROGEN REFINEMENT MODEL HAS BEEN USED WITH THE RIDING MODE
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 13.028 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.41→48.467 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj