+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6nyb | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a MAPK pathway complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Components Components |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | TRANSFERASE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationepithelial cell proliferation involved in lung morphogenesis / positive regulation of endodermal cell differentiation / negative regulation of homotypic cell-cell adhesion / negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway / regulation of vascular associated smooth muscle contraction / positive regulation of axon regeneration / CD4-positive, alpha-beta T cell differentiation / regulation of axon regeneration / mitogen-activated protein kinase kinase / CD4-positive or CD8-positive, alpha-beta T cell lineage commitment ...epithelial cell proliferation involved in lung morphogenesis / positive regulation of endodermal cell differentiation / negative regulation of homotypic cell-cell adhesion / negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway / regulation of vascular associated smooth muscle contraction / positive regulation of axon regeneration / CD4-positive, alpha-beta T cell differentiation / regulation of axon regeneration / mitogen-activated protein kinase kinase / CD4-positive or CD8-positive, alpha-beta T cell lineage commitment / negative regulation of synaptic vesicle exocytosis / positive regulation of muscle contraction / Golgi inheritance / placenta blood vessel development / MAP-kinase scaffold activity / Signalling to p38 via RIT and RIN / cerebellar cortex formation / labyrinthine layer development / melanosome transport / head morphogenesis / ARMS-mediated activation / type B pancreatic cell proliferation / endothelial cell apoptotic process / myeloid progenitor cell differentiation / Signaling by MAP2K mutants / SHOC2 M1731 mutant abolishes MRAS complex function / Gain-of-function MRAS complexes activate RAF signaling / negative regulation of fibroblast migration / positive regulation of D-glucose transmembrane transport / vesicle transport along microtubule / establishment of protein localization to membrane / positive regulation of axonogenesis / positive regulation of Ras protein signal transduction / regulation of Golgi inheritance / mitogen-activated protein kinase kinase kinase binding / central nervous system neuron differentiation / regulation of T cell differentiation / trachea formation / triglyceride homeostasis / Negative feedback regulation of MAPK pathway / regulation of early endosome to late endosome transport / regulation of stress-activated MAPK cascade / Frs2-mediated activation / stress fiber assembly / MAPK3 (ERK1) activation / ERBB2-ERBB3 signaling pathway / regulation of neurotransmitter receptor localization to postsynaptic specialization membrane / face development / endodermal cell differentiation / MAP kinase kinase activity / Bergmann glial cell differentiation / positive regulation of ATP biosynthetic process / thyroid gland development / Uptake and function of anthrax toxins / positive regulation of protein serine/threonine kinase activity / synaptic vesicle exocytosis / somatic stem cell population maintenance / positive regulation of peptidyl-serine phosphorylation / MAP kinase kinase kinase activity / protein kinase activator activity / negative regulation of endothelial cell apoptotic process / Schwann cell development / response to axon injury / postsynaptic modulation of chemical synaptic transmission / keratinocyte differentiation / neuron projection morphogenesis / positive regulation of stress fiber assembly / ERK1 and ERK2 cascade / myelination / positive regulation of substrate adhesion-dependent cell spreading / positive regulation of autophagy / protein serine/threonine/tyrosine kinase activity / dendrite cytoplasm / substrate adhesion-dependent cell spreading / insulin-like growth factor receptor signaling pathway / cellular response to calcium ion / response to glucocorticoid / thymus development / animal organ morphogenesis / MAP3K8 (TPL2)-dependent MAPK1/3 activation / protein serine/threonine kinase activator activity / Signal transduction by L1 / cell motility / positive regulation of transcription elongation by RNA polymerase II / RAF activation / Spry regulation of FGF signaling / Signaling by high-kinase activity BRAF mutants / MAP2K and MAPK activation / visual learning / small GTPase binding / cellular response to xenobiotic stimulus / epidermal growth factor receptor signaling pathway / centriolar satellite / long-term synaptic potentiation / chemotaxis / neuron differentiation / Negative regulation of MAPK pathway / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF Similarity search - Function | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) Spodoptera exigua (beet armyworm) Spodoptera exigua (beet armyworm) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Park, E. / Rawson, S. / Li, K. / Jeon, H. / Eck, M.J. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Funding support |  United States, 2items United States, 2items
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes. Authors: Eunyoung Park / Shaun Rawson / Kunhua Li / Byeong-Won Kim / Scott B Ficarro / Gonzalo Gonzalez-Del Pino / Humayun Sharif / Jarrod A Marto / Hyesung Jeon / Michael J Eck /  Abstract: RAF family kinases are RAS-activated switches that initiate signalling through the MAP kinase cascade to control cellular proliferation, differentiation and survival. RAF activity is tightly ...RAF family kinases are RAS-activated switches that initiate signalling through the MAP kinase cascade to control cellular proliferation, differentiation and survival. RAF activity is tightly regulated and inappropriate activation is a frequent cause of cancer; however, the structural basis for RAF regulation is poorly understood at present. Here we use cryo-electron microscopy to determine autoinhibited and active-state structures of full-length BRAF in complexes with MEK1 and a 14-3-3 dimer. The reconstruction reveals an inactive BRAF-MEK1 complex restrained in a cradle formed by the 14-3-3 dimer, which binds the phosphorylated S365 and S729 sites that flank the BRAF kinase domain. The BRAF cysteine-rich domain occupies a central position that stabilizes this assembly, but the adjacent RAS-binding domain is poorly ordered and peripheral. The 14-3-3 cradle maintains autoinhibition by sequestering the membrane-binding cysteine-rich domain and blocking dimerization of the BRAF kinase domain. In the active state, these inhibitory interactions are released and a single 14-3-3 dimer rearranges to bridge the C-terminal pS729 binding sites of two BRAFs, which drives the formation of an active, back-to-back BRAF dimer. Our structural snapshots provide a foundation for understanding normal RAF regulation and its mutational disruption in cancer and developmental syndromes. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6nyb.cif.gz 6nyb.cif.gz | 225.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6nyb.ent.gz pdb6nyb.ent.gz | 166.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6nyb.json.gz 6nyb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6nyb_validation.pdf.gz 6nyb_validation.pdf.gz | 1 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6nyb_full_validation.pdf.gz 6nyb_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  6nyb_validation.xml.gz 6nyb_validation.xml.gz | 39.1 KB | Display | |
| Data in CIF |  6nyb_validation.cif.gz 6nyb_validation.cif.gz | 59.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ny/6nyb https://data.pdbj.org/pub/pdb/validation_reports/ny/6nyb ftp://data.pdbj.org/pub/pdb/validation_reports/ny/6nyb ftp://data.pdbj.org/pub/pdb/validation_reports/ny/6nyb | HTTPS FTP |
-Related structure data
| Related structure data |  0541MC  6pp9C  6q0jC  6q0kC  6q0tC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 3 types, 4 molecules ABCD
| #1: Protein | Mass: 89402.789 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: BRAF, BRAF1, RAFB1 / Production host: Homo sapiens (human) / Gene: BRAF, BRAF1, RAFB1 / Production host:  References: UniProt: P15056, non-specific serine/threonine protein kinase |
|---|---|
| #2: Protein | Mass: 45934.543 Da / Num. of mol.: 1 / Mutation: S218A, S222A Source method: isolated from a genetically manipulated source Details: GDC-0623 / Source: (gene. exp.)  Homo sapiens (human) / Gene: MAP2K1, MEK1, PRKMK1 / Production host: Homo sapiens (human) / Gene: MAP2K1, MEK1, PRKMK1 / Production host:  References: UniProt: Q02750, mitogen-activated protein kinase kinase |
| #3: Protein | Mass: 28108.514 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Spodoptera exigua (beet armyworm) / References: UniProt: V9P4T4 Spodoptera exigua (beet armyworm) / References: UniProt: V9P4T4 |
-Non-polymers , 5 types, 6 molecules 



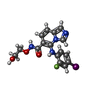




| #4: Chemical | ChemComp-AGS / | ||||||
|---|---|---|---|---|---|---|---|
| #5: Chemical | | #6: Chemical | ChemComp-ADP / | #7: Chemical | ChemComp-MG / | #8: Chemical | ChemComp-LCJ / | |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Ternary complex of BRAF/MEK1/14-3-3 with MEK inhibitor Type: COMPLEX Details: 14-3-3 is heterodimer of Insect epsilon and zeta. model was generated by Insect zeta sequence as a homo-dimer. Entity ID: #1-#3 / Source: MULTIPLE SOURCES |
|---|---|
| Molecular weight | Value: 0.19 MDa / Experimental value: YES |
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 7.4 |
| Specimen | Conc.: 0.05 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER |
| Electron lens | Mode: OTHER / Cs: 2.7 mm / C2 aperture diameter: 70 µm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.15.2_3472: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 4.1 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 165298 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller
















 PDBj
PDBj














