[English] 日本語
 Yorodumi
Yorodumi- PDB-4nxr: Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PD... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4nxr | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ Domain Quadruple Mutant (QM) in Complex With Neurexin-1 Peptide | ||||||
 Components Components |
| ||||||
 Keywords Keywords | signaling Protein/Peptide / Beta barrel fold protein / PDZ domain / peptide binding / specificity mutant / scaffold signaling protein for cell adhesion and cell junction / signaling domain / signaling Protein-Peptide complex | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of non-canonical Wnt signaling pathway / regulation of dopaminergic neuron differentiation / neuroligin family protein binding / Activated NTRK2 signals through CDK5 / neuron cell-cell adhesion / regulation of epithelial to mesenchymal transition / cell-cell contact zone / activation of GTPase activity / regulation of small GTPase mediated signal transduction / Wnt signaling pathway, planar cell polarity pathway ...regulation of non-canonical Wnt signaling pathway / regulation of dopaminergic neuron differentiation / neuroligin family protein binding / Activated NTRK2 signals through CDK5 / neuron cell-cell adhesion / regulation of epithelial to mesenchymal transition / cell-cell contact zone / activation of GTPase activity / regulation of small GTPase mediated signal transduction / Wnt signaling pathway, planar cell polarity pathway / Neurexins and neuroligins / positive regulation of axonogenesis / NRAGE signals death through JNK / small GTPase-mediated signal transduction / Rac protein signal transduction / CDC42 GTPase cycle / positive regulation of protein binding / EPH-ephrin mediated repulsion of cells / RHOA GTPase cycle / RAC2 GTPase cycle / RAC3 GTPase cycle / ephrin receptor signaling pathway / positive regulation of epithelial to mesenchymal transition / RAC1 GTPase cycle / EPHB-mediated forward signaling / cell adhesion molecule binding / guanyl-nucleotide exchange factor activity / cell projection / cell-matrix adhesion / kinase binding / cell-cell junction / transmembrane signaling receptor activity / cell migration / G alpha (12/13) signalling events / presynaptic membrane / protein-containing complex assembly / positive regulation of cell migration / positive regulation of cell population proliferation / synapse / lipid binding / signal transduction / metal ion binding / plasma membrane / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | ||||||
 Authors Authors | Liu, X. / Speckhard, D.C. / Shepherd, T.R. / Hengel, S.R. / Fuentes, E.J. | ||||||
 Citation Citation |  Journal: Structure / Year: 2016 Journal: Structure / Year: 2016Title: Distinct Roles for Conformational Dynamics in Protein-Ligand Interactions. Authors: Liu, X. / Speckhard, D.C. / Shepherd, T.R. / Sun, Y.J. / Hengel, S.R. / Yu, L. / Fowler, C.A. / Gakhar, L. / Fuentes, E.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4nxr.cif.gz 4nxr.cif.gz | 36.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4nxr.ent.gz pdb4nxr.ent.gz | 23.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4nxr.json.gz 4nxr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nx/4nxr https://data.pdbj.org/pub/pdb/validation_reports/nx/4nxr ftp://data.pdbj.org/pub/pdb/validation_reports/nx/4nxr ftp://data.pdbj.org/pub/pdb/validation_reports/nx/4nxr | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4nxpC  4nxqC  3kzdS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 10192.402 Da / Num. of mol.: 1 / Fragment: PDZ domain / Mutation: L911M, K912E, L915F, L920V Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Strain: human / Gene: TIAM1 / Plasmid: PET21A-6HIS-rTEV / Production host: Homo sapiens (human) / Strain: human / Gene: TIAM1 / Plasmid: PET21A-6HIS-rTEV / Production host:  |
|---|---|
| #2: Protein/peptide | Mass: 1060.158 Da / Num. of mol.: 1 / Fragment: UNP residue 659-666 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) / References: UniProt: P58401 Homo sapiens (human) / References: UniProt: P58401 |
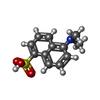
| #3: Chemical | ChemComp-EPE / |
| #4: Chemical | ChemComp-ANS / |
| #5: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.83 Å3/Da / Density % sol: 32.61 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 6.8 Details: 0.1M Sodium acetate, 0.1M HEPES, pH=7.5, 22% PEG 4000, pH 6.8, VAPOR DIFFUSION, HANGING DROP, temperature 291K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 4.2.2 / Wavelength: 1 Å / Beamline: 4.2.2 / Wavelength: 1 Å |
| Detector | Type: NOIR-1 / Detector: CCD / Date: Dec 14, 2012 |
| Radiation | Monochromator: double crystal Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→39.06 Å / Num. obs: 6821 / % possible obs: 98.7 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 6.7 % / Biso Wilson estimate: 11.4 Å2 / Rmerge(I) obs: 0.033 / Net I/σ(I): 39.7 |
| Reflection shell | Resolution: 1.9→1.94 Å / Redundancy: 4.4 % / Rmerge(I) obs: 0.035 / Mean I/σ(I) obs: 11 / % possible all: 87.1 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 3KZD Resolution: 1.9→39.06 Å / SU ML: 0.15 / σ(F): 0.64 / Phase error: 18.87 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→39.06 Å
| |||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj