[English] 日本語
 Yorodumi
Yorodumi- PDB-3zrm: Identification of 2-(4-pyridyl)thienopyridinones as GSK-3beta inh... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3zrm | ||||||
|---|---|---|---|---|---|---|---|
| Title | Identification of 2-(4-pyridyl)thienopyridinones as GSK-3beta inhibitors | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSFERASE/PEPTIDE / TRANSFERASE-PEPTIDE COMPLEX / KINASE | ||||||
| Function / homology |  Function and homology information Function and homology informationneuron projection organization / regulation of microtubule anchoring at centrosome / negative regulation of type B pancreatic cell development / negative regulation of glycogen (starch) synthase activity / negative regulation of mesenchymal stem cell differentiation / beta-arrestin-dependent dopamine receptor signaling pathway / superior temporal gyrus development / positive regulation of protein localization to cilium / negative regulation of glycogen biosynthetic process / negative regulation of TORC2 signaling ...neuron projection organization / regulation of microtubule anchoring at centrosome / negative regulation of type B pancreatic cell development / negative regulation of glycogen (starch) synthase activity / negative regulation of mesenchymal stem cell differentiation / beta-arrestin-dependent dopamine receptor signaling pathway / superior temporal gyrus development / positive regulation of protein localization to cilium / negative regulation of glycogen biosynthetic process / negative regulation of TORC2 signaling / negative regulation of dopaminergic neuron differentiation / maintenance of cell polarity / positive regulation of protein localization to centrosome / positive regulation of cilium assembly / CRMPs in Sema3A signaling / heart valve development / tau-protein kinase / beta-catenin destruction complex / APC truncation mutants have impaired AXIN binding / AXIN missense mutants destabilize the destruction complex / Truncations of AMER1 destabilize the destruction complex / regulation of protein export from nucleus / positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway / Maturation of nucleoprotein / cellular response to interleukin-3 / Beta-catenin phosphorylation cascade / Signaling by GSK3beta mutants / CTNNB1 S33 mutants aren't phosphorylated / CTNNB1 S37 mutants aren't phosphorylated / CTNNB1 S45 mutants aren't phosphorylated / CTNNB1 T41 mutants aren't phosphorylated / regulation of long-term synaptic potentiation / Wnt signalosome / negative regulation of TOR signaling / regulation of microtubule-based process / negative regulation of protein localization to nucleus / AKT phosphorylates targets in the cytosol / Disassembly of the destruction complex and recruitment of AXIN to the membrane / negative regulation of calcineurin-NFAT signaling cascade / regulation of axon extension / Maturation of nucleoprotein / tau-protein kinase activity / molecular function inhibitor activity / negative regulation of epithelial to mesenchymal transition / positive regulation of cell-matrix adhesion / ER overload response / regulation of axonogenesis / glycogen metabolic process / regulation of dendrite morphogenesis / regulation of neuron projection development / Constitutive Signaling by AKT1 E17K in Cancer / protein kinase A catalytic subunit binding / establishment of cell polarity / dynactin binding / epithelial to mesenchymal transition / positive regulation of protein binding / Regulation of HSF1-mediated heat shock response / canonical Wnt signaling pathway / NF-kappaB binding / negative regulation of osteoblast differentiation / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / negative regulation of protein-containing complex assembly / regulation of cellular response to heat / extrinsic apoptotic signaling pathway / cellular response to retinoic acid / positive regulation of type I interferon production / extrinsic apoptotic signaling pathway in absence of ligand / positive regulation of autophagy / Transcriptional and post-translational regulation of MITF-M expression and activity / presynaptic modulation of chemical synaptic transmission / regulation of microtubule cytoskeleton organization / negative regulation of cell migration / response to endoplasmic reticulum stress / positive regulation of protein ubiquitination / positive regulation of protein export from nucleus / hippocampus development / mitochondrion organization / Ubiquitin-dependent degradation of Cyclin D / excitatory postsynaptic potential / positive regulation of cell differentiation / peptidyl-serine phosphorylation / positive regulation of protein-containing complex assembly / negative regulation of canonical Wnt signaling pathway / circadian rhythm / GSK3B and BTRC:CUL1-mediated-degradation of NFE2L2 / regulation of circadian rhythm / Degradation of GLI2 by the proteasome / GLI3 is processed to GLI3R by the proteasome / beta-catenin binding / Degradation of beta-catenin by the destruction complex / B-WICH complex positively regulates rRNA expression / tau protein binding / cellular response to amyloid-beta / Wnt signaling pathway / neuron projection development / Regulation of RUNX2 expression and activity / p53 binding / kinase activity / positive regulation of protein catabolic process / insulin receptor signaling pathway Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  FOURIER SYNTHESIS / Resolution: 2.49 Å FOURIER SYNTHESIS / Resolution: 2.49 Å | ||||||
 Authors Authors | Gentile, G. / Bernasconi, G. / Pozzan, A. / Merlo, G. / Marzorati, P. / Bamborough, P. / Bax, B. / Bridges, A. / Brough, C. / Carter, P. ...Gentile, G. / Bernasconi, G. / Pozzan, A. / Merlo, G. / Marzorati, P. / Bamborough, P. / Bax, B. / Bridges, A. / Brough, C. / Carter, P. / Cutler, G. / Neu, M. / Takada, M. | ||||||
 Citation Citation |  Journal: Bioorg.Med.Chem.Lett. / Year: 2011 Journal: Bioorg.Med.Chem.Lett. / Year: 2011Title: Identification of 2-(4-Pyridyl)Thienopyridinones as Gsk-3Beta Inhibitors. Authors: Gentile, G. / Bernasconi, G. / Pozzan, A. / Merlo, G. / Marzorati, P. / Bamborough, P. / Bax, B. / Bridges, A. / Brough, C. / Carter, P. / Cutler, G. / Neu, M. / Takada, M. | ||||||
| History |
| ||||||
| Remark 700 | SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "AA" IN EACH CHAIN ON SHEET RECORDS BELOW ... SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "AA" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 6-STRANDED BARREL THIS IS REPRESENTED BY A 7-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. THE SHEETS PRESENTED AS "BA" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 6-STRANDED BARREL THIS IS REPRESENTED BY A 7-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3zrm.cif.gz 3zrm.cif.gz | 167.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3zrm.ent.gz pdb3zrm.ent.gz | 133.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3zrm.json.gz 3zrm.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zr/3zrm https://data.pdbj.org/pub/pdb/validation_reports/zr/3zrm ftp://data.pdbj.org/pub/pdb/validation_reports/zr/3zrm ftp://data.pdbj.org/pub/pdb/validation_reports/zr/3zrm | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3zrkC  3zrlC  1gngS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein / Protein/peptide , 2 types, 4 molecules ABXY
| #1: Protein | Mass: 42036.117 Da / Num. of mol.: 2 / Fragment: RESIDUES 23-393 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Cell line (production host): SF9 / Production host: HOMO SAPIENS (human) / Cell line (production host): SF9 / Production host:  #2: Protein/peptide | Mass: 3772.434 Da / Num. of mol.: 2 / Fragment: RESIDUES 197-226 Source method: isolated from a genetically manipulated source Details: FRATTIDE SEQUENCE CO-EXPRESSED IN BACULOVIRUS (DUAL EXPRESSION) Source: (gene. exp.)  HOMO SAPIENS (human) / Cell line (production host): SF9 / Production host: HOMO SAPIENS (human) / Cell line (production host): SF9 / Production host:  |
|---|
-Non-polymers , 4 types, 193 molecules 

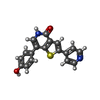




| #3: Chemical | ChemComp-SO4 / #4: Chemical | #5: Chemical | #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.56 Å3/Da / Density % sol: 51.65 % / Description: NONE |
|---|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID23-1 / Wavelength: 1.0723 / Beamline: ID23-1 / Wavelength: 1.0723 |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.0723 Å / Relative weight: 1 |
| Reflection | Resolution: 2.49→40 Å / Num. obs: 31863 / % possible obs: 100 % / Observed criterion σ(I): 0 / Redundancy: 7.4 % / Rmerge(I) obs: 0.1 / Net I/σ(I): 24.2 |
| Reflection shell | Resolution: 2.49→2.53 Å / Redundancy: 7.4 % / Rmerge(I) obs: 0.49 / Mean I/σ(I) obs: 5.1 / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  FOURIER SYNTHESIS FOURIER SYNTHESISStarting model: PDB ENTRY 1GNG Resolution: 2.49→20 Å / Cor.coef. Fo:Fc: 0.947 / Cor.coef. Fo:Fc free: 0.911 / SU B: 8.049 / SU ML: 0.184 / Cross valid method: THROUGHOUT / ESU R: 0.52 / ESU R Free: 0.283 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 41.067 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.49→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller

































 PDBj
PDBj








