[English] 日本語
 Yorodumi
Yorodumi- PDB-3r1z: Crystal structure of NYSGRC enolase target 200555, a putative dip... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3r1z | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of NYSGRC enolase target 200555, a putative dipeptide epimerase from Francisella philomiragia : Complex with L-Ala-L-Glu and L-Ala-D-Glu | ||||||
 Components Components | Enzyme of enolase superfamily | ||||||
 Keywords Keywords | ISOMERASE / enolase / structural genomics / putative epimerase / PSI-Biology / New York Structural Genomics Research Consortium / NYSGRC | ||||||
| Function / homology |  Function and homology information Function and homology informationracemase and epimerase activity, acting on amino acids and derivatives / Isomerases; Racemases and epimerases; Acting on amino acids and derivatives / racemase and epimerase activity / peptide metabolic process / magnesium ion binding Similarity search - Function | ||||||
| Biological species |  Francisella philomiragia subsp. philomiragia (bacteria) Francisella philomiragia subsp. philomiragia (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  FOURIER SYNTHESIS / Resolution: 1.9 Å FOURIER SYNTHESIS / Resolution: 1.9 Å | ||||||
 Authors Authors | Vetting, M.W. / Hillerich, B. / Seidel, R.D. / Zencheck, W.D. / Toro, R. / Imker, H.J. / Gerlt, J.A. / Almo, S.C. / New York Structural Genomics Research Consortium (NYSGRC) | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2012 Journal: Proc.Natl.Acad.Sci.USA / Year: 2012Title: Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily. Authors: Lukk, T. / Sakai, A. / Kalyanaraman, C. / Brown, S.D. / Imker, H.J. / Song, L. / Fedorov, A.A. / Fedorov, E.V. / Toro, R. / Hillerich, B. / Seidel, R. / Patskovsky, Y. / Vetting, M.W. / ...Authors: Lukk, T. / Sakai, A. / Kalyanaraman, C. / Brown, S.D. / Imker, H.J. / Song, L. / Fedorov, A.A. / Fedorov, E.V. / Toro, R. / Hillerich, B. / Seidel, R. / Patskovsky, Y. / Vetting, M.W. / Nair, S.K. / Babbitt, P.C. / Almo, S.C. / Gerlt, J.A. / Jacobson, M.P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3r1z.cif.gz 3r1z.cif.gz | 302.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3r1z.ent.gz pdb3r1z.ent.gz | 246.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3r1z.json.gz 3r1z.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/r1/3r1z https://data.pdbj.org/pub/pdb/validation_reports/r1/3r1z ftp://data.pdbj.org/pub/pdb/validation_reports/r1/3r1z ftp://data.pdbj.org/pub/pdb/validation_reports/r1/3r1z | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3ijiC  3ijlC  3ijqC  3ik4C  3jvaC  3jw7C  3jzuC  3k1gC  3kumC  3q45C  3q4dC  3r0kC  3r0uC  3r10C  3r11C  3ritC  3ro6C C: citing same article ( |
|---|---|
| Similar structure data | |
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 42455.102 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Francisella philomiragia subsp. philomiragia (bacteria) Francisella philomiragia subsp. philomiragia (bacteria)Strain: ATCC 25017 / Gene: Fphi_1647 / Plasmid: CHS30 / Production host:  |
|---|
-Non-polymers , 6 types, 722 molecules 



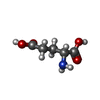






| #2: Chemical | | #3: Chemical | ChemComp-GLU / | #4: Chemical | ChemComp-SO4 / #5: Chemical | ChemComp-GOL / #6: Chemical | ChemComp-DGL / | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.22 Å3/Da / Density % sol: 61.82 % |
|---|---|
| Crystal grow | Temperature: 294 K / Method: vapor diffusion, sitting drop / pH: 5.6 Details: Protein: 10 mM HEPES pH 7.5, 150 mM Nacl, 10% glycerol, 5 mM DTT; Reservoir: 2M Ammonium Sulfate, 100 mM NaCitrate, 200 mM KNaTartrate; Soak: 2.2 M Ammonium sulfate, 100 mM MES pH 6.0, 18% ...Details: Protein: 10 mM HEPES pH 7.5, 150 mM Nacl, 10% glycerol, 5 mM DTT; Reservoir: 2M Ammonium Sulfate, 100 mM NaCitrate, 200 mM KNaTartrate; Soak: 2.2 M Ammonium sulfate, 100 mM MES pH 6.0, 18% glycerol, 50mM MgSO4, 25 mM L-Ala-L-Glu, 30 min, vapour diffusion, sitting drop, temperature 294K |
-Data collection
| Diffraction | Mean temperature: 98 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 31-ID / Wavelength: 0.97905 Å / Beamline: 31-ID / Wavelength: 0.97905 Å |
| Detector | Type: RAYONIX MX225HE / Detector: CCD / Details: Undulator Source |
| Radiation | Monochromator: diamond / Protocol: SINGLE WAVELENGTH / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97905 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→50 Å / Num. all: 85703 / Num. obs: 85703 / % possible obs: 97.9 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 12.5 % / Biso Wilson estimate: 19.31 Å2 / Rmerge(I) obs: 0.097 / Rsym value: 0.097 / Net I/σ(I): 16.4 |
| Reflection shell | Resolution: 1.9→2 Å / Redundancy: 12.1 % / Rmerge(I) obs: 0.38 / Mean I/σ(I) obs: 5.8 / Num. unique all: 3006 / Rsym value: 0.38 / % possible all: 92.5 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  FOURIER SYNTHESIS / Resolution: 1.9→29.69 Å / Occupancy max: 1 / Occupancy min: 0.21 / FOM work R set: 0.8984 / SU ML: 0.18 / σ(F): 0 / σ(I): 0 / Phase error: 16.75 / Stereochemistry target values: ML FOURIER SYNTHESIS / Resolution: 1.9→29.69 Å / Occupancy max: 1 / Occupancy min: 0.21 / FOM work R set: 0.8984 / SU ML: 0.18 / σ(F): 0 / σ(I): 0 / Phase error: 16.75 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.95 Å / VDW probe radii: 1.2 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 43.043 Å2 / ksol: 0.378 e/Å3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 98.02 Å2 / Biso mean: 24.6422 Å2 / Biso min: 5.25 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→29.69 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 10
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj







