[English] 日本語
 Yorodumi
Yorodumi- PDB-3est: STRUCTURE OF NATIVE PORCINE PANCREATIC ELASTASE AT 1.65 ANGSTROMS... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3est | ||||||
|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF NATIVE PORCINE PANCREATIC ELASTASE AT 1.65 ANGSTROMS RESOLUTION | ||||||
 Components Components | PORCINE PANCREATIC ELASTASE | ||||||
 Keywords Keywords | HYDROLASE(SERINE PROTEINASE) | ||||||
| Function / homology |  Function and homology information Function and homology informationpancreatic elastase / serine-type endopeptidase activity / proteolysis / extracellular space / metal ion binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.65 Å X-RAY DIFFRACTION / Resolution: 1.65 Å | ||||||
 Authors Authors | Meyer, E.F. / Cole, G. / Radhakrishnan, R. / Epp, O. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.B / Year: 1988 Journal: Acta Crystallogr.,Sect.B / Year: 1988Title: Structure of native porcine pancreatic elastase at 1.65 A resolutions. Authors: Meyer, E. / Cole, G. / Radhakrishnan, R. / Epp, O. #1:  Journal: J.Mol.Biol. / Year: 1987 Journal: J.Mol.Biol. / Year: 1987Title: Crystal Structures of the Complex of Porcine Pancreatic Elastase with Two Valine-Derived Benzoxazinone Inhibitors Authors: Radhakrishnan, R. / Presta, L.G. / Meyerjunior, E.F. / Wildonger, R. #2:  Journal: Biochemistry / Year: 1988 Journal: Biochemistry / Year: 1988Title: Analysis of an Enzyme-Substrate Complex by X-Ray Crystallography and Transferred Nuclear Overhauser Enhancement Measurements: Porcine Pancreatic Elastase and a Hexapeptide Authors: Meyerjunior, E.F. / Clore, G.M. / Gronenborn, A.M. / Hansen, H.A.S. #3:  Journal: J.Mol.Biol. / Year: 1986 Journal: J.Mol.Biol. / Year: 1986Title: Structure of the Product Complex of Acetyl-Ala-Pro-Ala with Porcine Pancreatic Elastase at 1.65 Angstroms Resolution Authors: Meyerjunior, E.F. / Radhakrishnan, R. / Cole, G.M. / Presta, L.G. #4:  Journal: J.Mol.Biol. / Year: 1986 Journal: J.Mol.Biol. / Year: 1986Title: Stereochemistry of Binding of the Tetrapeptide Acetyl-Pro-Ala-Pro-Tyr-NH2 to Porcine Pancreatic Elastase. Combined Use of Two-Dimensional Transferred Nuclear Overhauser Enhancement ...Title: Stereochemistry of Binding of the Tetrapeptide Acetyl-Pro-Ala-Pro-Tyr-NH2 to Porcine Pancreatic Elastase. Combined Use of Two-Dimensional Transferred Nuclear Overhauser Enhancement Measurements, Restrained Molecular Dynamics, X-Ray Crystallography and Molecular Modelling Authors: Clore, G.M. / Gronenborn, A.M. / Carlson, G. / Meyer, E.F. #5:  Journal: J.Am.Chem.Soc. / Year: 1985 Journal: J.Am.Chem.Soc. / Year: 1985Title: Stereospecific Reaction of 3-Methoxy-4-Chloro-7-Amino-Isocoumarin with Crystalline Porcine Pancreatic Elastase Authors: Meyerjunior, E.F. / Presta, L.G. / Radhakrishnan, R. #6:  Journal: J.Mol.Biol. / Year: 1978 Journal: J.Mol.Biol. / Year: 1978Title: The Atomic Structure of Crystalline Porcine Pancreatic Elastase at 2.5 Angstroms Resolution. Comparisons with the Structure of Alpha-Chymotrypsin Authors: Sawyer, L. / Shotton, D.M. / Campbell, J.W. / Wendell, P.L. / Muirhead, H. / Watson, H.C. / Diamond, R. / Ladner, R.C. #7:  Journal: Biochem.Biophys.Res.Commun. / Year: 1973 Journal: Biochem.Biophys.Res.Commun. / Year: 1973Title: Atomic Coordinates for Tosyl-Elastase Authors: Sawyer, L. / Shotton, D.M. / Watson, H.C. #8:  Journal: Nature / Year: 1970 Journal: Nature / Year: 1970Title: Amino-Acid Sequence of Porcine Pancreatic Elastase and its Homologies with Other Serine Proteinases Authors: Shotton, D.M. / Hartley, B.S. #9:  Journal: Nature / Year: 1970 Journal: Nature / Year: 1970Title: Three-Dimensional Fourier Synthesis of Tosyl-Elastase at 3.5 Angstroms Resolution Authors: Watson, H.C. / Shotton, D.M. / Cox, J.M. / Muirhead, H. #10:  Journal: Nature / Year: 1970 Journal: Nature / Year: 1970Title: Three Dimensional Structure of Tosyl-Elastase Authors: Shotton, D.M. / Watson, H.C. | ||||||
| History |
| ||||||
| Remark 700 | SHEET THE TWO SEVEN STRANDED SHEETS IN THIS STRUCTURE ARE REALLY SIX STRANDED BETA BARRELS. THIS IS ...SHEET THE TWO SEVEN STRANDED SHEETS IN THIS STRUCTURE ARE REALLY SIX STRANDED BETA BARRELS. THIS IS DENOTED BY THE FIRST STRAND RECURRING AS THE LAST STRAND. |
- Structure visualization
Structure visualization
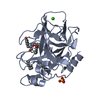
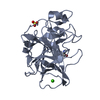
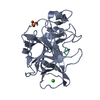
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3est.cif.gz 3est.cif.gz | 63 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3est.ent.gz pdb3est.ent.gz | 45.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3est.json.gz 3est.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  3est_validation.pdf.gz 3est_validation.pdf.gz | 427.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  3est_full_validation.pdf.gz 3est_full_validation.pdf.gz | 432 KB | Display | |
| Data in XML |  3est_validation.xml.gz 3est_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  3est_validation.cif.gz 3est_validation.cif.gz | 17.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/es/3est https://data.pdbj.org/pub/pdb/validation_reports/es/3est ftp://data.pdbj.org/pub/pdb/validation_reports/es/3est ftp://data.pdbj.org/pub/pdb/validation_reports/es/3est | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: THE FOLLOWING ATOMS WERE TREATED AS DUMMY ATOMS, WHICH MEANS THAT THEY CONTRIBUTED TO THE ENERGY REFINEMENT PART OF EREF BUT NOT TO THE CALCULATED STRUCTURE FACTORS -SER 36C OG, SER 37 OG, ARG 61 ...1: THE FOLLOWING ATOMS WERE TREATED AS DUMMY ATOMS, WHICH MEANS THAT THEY CONTRIBUTED TO THE ENERGY REFINEMENT PART OF EREF BUT NOT TO THE CALCULATED STRUCTURE FACTORS -SER 36C OG, SER 37 OG, ARG 61 (CZ,NH1,NH2), GLN 110 (CG,CD,OE1,NE2), ARG 125 (CG,CD,NE,CZ,NH1,NH2), SO4 290 (O1,O3). |
- Components
Components
| #1: Protein | Mass: 25928.031 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  | ||||||
|---|---|---|---|---|---|---|---|
| #2: Chemical | ChemComp-CA / | ||||||
| #3: Chemical | | #4: Water | ChemComp-HOH / | Has protein modification | Y | Nonpolymer details | SEVEN GROUPS OF WATER MOLECULES ARE REPORTED. THESE TEND TO BE CONSERVED IN THE FAMILY OF SERINE ...SEVEN GROUPS OF WATER MOLECULES ARE REPORTED. THESE TEND TO BE CONSERVED IN THE FAMILY OF SERINE PROTEASES. FUNCTIONAL | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.19 Å3/Da / Density % sol: 43.94 % | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Method: microdialysisDetails: took Meyer, Radhakrishman et al., (1986) from original paper | |||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Reflection | *PLUS Highest resolution: 1.65 Å / Num. obs: 19232 / % possible obs: 66 % / Rmerge(I) obs: 0.093 |
|---|
- Processing
Processing
| Software | Name: EREF / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Rfactor Rwork: 0.169 / Highest resolution: 1.65 Å Details: THE FOLLOWING ATOMS WERE TREATED AS DUMMY ATOMS, WHICH MEANS THAT THEY CONTRIBUTED TO THE ENERGY REFINEMENT PART OF EREF BUT NOT TO THE CALCULATED STRUCTURE FACTORS -SER 36C OG, SER 37 OG, ...Details: THE FOLLOWING ATOMS WERE TREATED AS DUMMY ATOMS, WHICH MEANS THAT THEY CONTRIBUTED TO THE ENERGY REFINEMENT PART OF EREF BUT NOT TO THE CALCULATED STRUCTURE FACTORS -SER 36C OG, SER 37 OG, ARG 61 (CZ,NH1,NH2), GLN 110 (CG,CD,OE1,NE2), ARG 125 (CG,CD,NE,CZ,NH1,NH2), SO4 290 (O1,O3). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 1.65 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Lowest resolution: 7 Å / Rfactor all: 0.169 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 14.1 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller



















 PDBj
PDBj





