[English] 日本語
 Yorodumi
Yorodumi- PDB-2ygv: Conserved N-terminal domain of the yeast Histone Chaperone Asf1 i... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2ygv | ||||||
|---|---|---|---|---|---|---|---|
| Title | Conserved N-terminal domain of the yeast Histone Chaperone Asf1 in complex with the C-terminal fragment of Rad53 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | CHAPERONE/TRANSFERASE / CHAPERONE-TRANSFERASE COMPLEX / CHECKPOINT / DNA DAMAGE / CHROMATIN | ||||||
| Function / homology |  Function and homology information Function and homology informationdeoxyribonucleoside triphosphate biosynthetic process / Formation of Senescence-Associated Heterochromatin Foci (SAHF) / H3 histone acetyltransferase complex / meiotic recombination checkpoint signaling / DNA replication-dependent chromatin assembly / acetyltransferase activator activity / dual-specificity kinase / nucleosome disassembly / silent mating-type cassette heterochromatin formation / cellular response to stress ...deoxyribonucleoside triphosphate biosynthetic process / Formation of Senescence-Associated Heterochromatin Foci (SAHF) / H3 histone acetyltransferase complex / meiotic recombination checkpoint signaling / DNA replication-dependent chromatin assembly / acetyltransferase activator activity / dual-specificity kinase / nucleosome disassembly / silent mating-type cassette heterochromatin formation / cellular response to stress / telomere maintenance in response to DNA damage / negative regulation of DNA damage checkpoint / DNA replication origin binding / DNA replication initiation / subtelomeric heterochromatin formation / regulation of DNA repair / protein serine/threonine/tyrosine kinase activity / DNA damage checkpoint signaling / protein modification process / positive regulation of transcription elongation by RNA polymerase II / intracellular protein localization / nucleosome assembly / chromatin organization / regulation of gene expression / protein tyrosine kinase activity / histone binding / chromosome, telomeric region / protein kinase activity / protein serine kinase activity / DNA repair / protein serine/threonine kinase activity / regulation of transcription by RNA polymerase II / chromatin / ATP binding / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.94 Å MOLECULAR REPLACEMENT / Resolution: 2.94 Å | ||||||
 Authors Authors | Jiao, Y. / Seeger, K. / Murciano, B. / Ledu, M.H. / Charbonnier, J.B. / Legrand, P. / Lautrette, A. / Gaubert, A. / Mousson, F. / Guerois, R. ...Jiao, Y. / Seeger, K. / Murciano, B. / Ledu, M.H. / Charbonnier, J.B. / Legrand, P. / Lautrette, A. / Gaubert, A. / Mousson, F. / Guerois, R. / Mann, C. / Ochsenbein, F. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2012 Journal: Proc.Natl.Acad.Sci.USA / Year: 2012Title: Surprising Complexity of the Asf1 Histone Chaperone-Rad53 Kinase Interaction Authors: Jiao, Y. / Seeger, K. / Lautrette, A. / Gaubert, A. / Mousson, F. / Guerois, R. / Mann, C. / Ochsenbein, F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2ygv.cif.gz 2ygv.cif.gz | 145.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2ygv.ent.gz pdb2ygv.ent.gz | 116 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2ygv.json.gz 2ygv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yg/2ygv https://data.pdbj.org/pub/pdb/validation_reports/yg/2ygv ftp://data.pdbj.org/pub/pdb/validation_reports/yg/2ygv ftp://data.pdbj.org/pub/pdb/validation_reports/yg/2ygv | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1rocS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 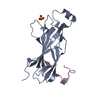
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 17874.141 Da / Num. of mol.: 4 / Fragment: CONSERVED N-TERMINAL DOMAIN, RESIDUES 1-156 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Strain: W303 / Production host:  #2: Protein/peptide | Mass: 2495.872 Da / Num. of mol.: 4 / Fragment: C-TERMINAL, RESIDUES 800-821 / Source method: obtained synthetically / Source: (synth.)  References: UniProt: P22216, non-specific serine/threonine protein kinase #3: Chemical | ChemComp-SO4 / #4: Chemical | ChemComp-GOL / | #5: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.41 Å3/Da / Density % sol: 48.88 % / Description: NONE |
|---|---|
| Crystal grow | pH: 6 / Details: 5% PEG 8000, 0.1M NACACO PH 6, 0.2M MG ACETATE |
-Data collection
| Diffraction | Mean temperature: 287 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SOLEIL SOLEIL  / Beamline: PROXIMA 1 / Wavelength: 0.9 / Beamline: PROXIMA 1 / Wavelength: 0.9 |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Nov 8, 2009 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9 Å / Relative weight: 1 |
| Reflection | Resolution: 2.94→38.9 Å / Num. obs: 16378 / % possible obs: 98.7 % / Observed criterion σ(I): 2.36 / Redundancy: 3.96 % / Biso Wilson estimate: 61.41 Å2 / Rmerge(I) obs: 0.02 / Net I/σ(I): 10.59 |
| Reflection shell | Resolution: 2.94→3.12 Å / Redundancy: 3.82 % / Rmerge(I) obs: 0.09 / Mean I/σ(I) obs: 2.36 / % possible all: 97.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1ROC Resolution: 2.94→38.9 Å / Cor.coef. Fo:Fc: 0.867 / Cor.coef. Fo:Fc free: 0.8082 / Cross valid method: THROUGHOUT / σ(F): 0 Details: IDEAL-DIST CONTACT TERM CONTACT SETUP. ALL ATOMS HAVE CCP4 ATOM TYPE FROM LIBRARY.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 54.12 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.367 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.94→38.9 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.94→3.14 Å / Total num. of bins used: 8
|
 Movie
Movie Controller
Controller





























 PDBj
PDBj







