+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2byk | ||||||
|---|---|---|---|---|---|---|---|
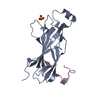
| Title | Histone fold heterodimer of the Chromatin Accessibility Complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA BINDING PROTEIN / CHRAC-14 / NUCLEOSOME SLIDING / HISTONE FOLD / CHRAC-16 / DNA-BINDING PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationDNA replication initiation / PCNA-Dependent Long Patch Base Excision Repair / Termination of translesion DNA synthesis / Recognition of DNA damage by PCNA-containing replication complex / Dual Incision in GG-NER / Dual incision in TC-NER / Activation of the pre-replicative complex / CHRAC / epsilon DNA polymerase complex / CENP-A containing chromatin assembly ...DNA replication initiation / PCNA-Dependent Long Patch Base Excision Repair / Termination of translesion DNA synthesis / Recognition of DNA damage by PCNA-containing replication complex / Dual Incision in GG-NER / Dual incision in TC-NER / Activation of the pre-replicative complex / CHRAC / epsilon DNA polymerase complex / CENP-A containing chromatin assembly / ATAC complex / leading strand elongation / cellular response to gamma radiation / chromatin DNA binding / DNA-templated DNA replication / heterochromatin formation / nucleic acid binding / molecular adaptor activity / chromatin remodeling / protein heterodimerization activity / DNA damage response / nucleus Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.4 Å MOLECULAR REPLACEMENT / Resolution: 2.4 Å | ||||||
 Authors Authors | Fernandez-Tornero, C. / Hartlepp, K.F. / Grune, T. / Eberharter, A. / Becker, P.B. / Muller, C.W. | ||||||
 Citation Citation |  Journal: Mol.Cell.Biol. / Year: 2005 Journal: Mol.Cell.Biol. / Year: 2005Title: The Histone Fold Subunits of Drosophila Chrac Facilitate Nucleosome Sliding Through Dynamic DNA Interactions. Authors: Hartlepp, K.F. / Fernandez-Tornero, C. / Eberharter, A. / Grune, T. / Muller, C.W. / Becker, P.B. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2byk.cif.gz 2byk.cif.gz | 77.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2byk.ent.gz pdb2byk.ent.gz | 57 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2byk.json.gz 2byk.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/by/2byk https://data.pdbj.org/pub/pdb/validation_reports/by/2byk ftp://data.pdbj.org/pub/pdb/validation_reports/by/2byk ftp://data.pdbj.org/pub/pdb/validation_reports/by/2byk | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2bymSC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
| #1: Protein | Mass: 16020.667 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Protein | Mass: 13862.586 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #3: Chemical | #4: Water | ChemComp-HOH / | Sequence details | PROTEOLYTIC CLEAVAGE IN THE CRYSTALLIZATION DROP MIGHT HAVE OCCURRED BUT ATTEMPTS TO CHARACTERIZE ...PROTEOLYTI | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.31 Å3/Da / Density % sol: 46.3 % |
|---|---|
| Crystal grow | pH: 3.5 / Details: 2M AMMONIUM SULFATE, 0.1M CITRIC ACID, PH 3.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-4 / Wavelength: 0.9393 / Beamline: ID14-4 / Wavelength: 0.9393 |
| Detector | Type: ADSC CCD / Detector: CCD |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9393 Å / Relative weight: 1 |
| Reflection | Resolution: 2.4→24 Å / Num. obs: 22490 / % possible obs: 99.9 % / Observed criterion σ(I): 2 / Redundancy: 5.8 % / Biso Wilson estimate: 51.6 Å2 / Rmerge(I) obs: 0.07 / Net I/σ(I): 4.4 |
| Reflection shell | Resolution: 2.4→2.5 Å / Redundancy: 5.7 % / Rmerge(I) obs: 0.26 / Mean I/σ(I) obs: 3 / % possible all: 99.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2BYM Resolution: 2.4→23.83 Å / Rfactor Rfree error: 0.006 / Data cutoff high absF: 1896010.14 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 73.8532 Å2 / ksol: 0.388442 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 66 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.4→23.83 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.4→2.55 Å / Rfactor Rfree error: 0.017 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj



