[English] 日本語
 Yorodumi
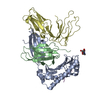
Yorodumi- PDB-2dyp: Crystal Structure of LILRB2(LIR2/ILT4/CD85d) complexed with HLA-G -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2dyp | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of LILRB2(LIR2/ILT4/CD85d) complexed with HLA-G | ||||||
 Components Components |
| ||||||
 Keywords Keywords | IMMUNE SYSTEM / Immunoglobulin-like | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of antigen processing and presentation / negative regulation of T cell costimulation / peripheral B cell tolerance induction / positive regulation of tolerance induction / MHC class Ib protein complex binding / immune response-inhibiting cell surface receptor signaling pathway / inhibitory MHC class I receptor activity / negative regulation of dendritic cell differentiation / Fc receptor mediated inhibitory signaling pathway / negative regulation of postsynaptic density organization ...negative regulation of antigen processing and presentation / negative regulation of T cell costimulation / peripheral B cell tolerance induction / positive regulation of tolerance induction / MHC class Ib protein complex binding / immune response-inhibiting cell surface receptor signaling pathway / inhibitory MHC class I receptor activity / negative regulation of dendritic cell differentiation / Fc receptor mediated inhibitory signaling pathway / negative regulation of postsynaptic density organization / positive regulation of natural killer cell cytokine production / positive regulation of long-term synaptic depression / MHC class Ib protein binding / negative regulation of T cell mediated cytotoxicity / immune response-regulating signaling pathway / cis-Golgi network membrane / negative regulation of immune response / positive regulation of T cell tolerance induction / interleukin-10-mediated signaling pathway / protein phosphatase 1 binding / negative regulation of G0 to G1 transition / regulation of dendritic cell differentiation / XY body / protein localization to site of double-strand break / chromatin-protein adaptor activity / negative regulation of natural killer cell mediated cytotoxicity / filopodium membrane / regulation of long-term synaptic potentiation / negative regulation of protein metabolic process / heterotypic cell-cell adhesion / positive regulation of regulatory T cell differentiation / positive regulation of macrophage cytokine production / response to ionizing radiation / CD8 receptor binding / protein homotrimerization / negative regulation of calcium ion transport / MHC class I protein binding / protection from natural killer cell mediated cytotoxicity / tertiary granule membrane / site of DNA damage / ficolin-1-rich granule membrane / positive regulation of endothelial cell apoptotic process / antigen processing and presentation of endogenous peptide antigen via MHC class Ib / antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-independent / cellular defense response / negative regulation of T cell proliferation / cell adhesion molecule binding / Replacement of protamines by nucleosomes in the male pronucleus / Packaging Of Telomere Ends / positive regulation of interleukin-12 production / positive regulation of T cell proliferation / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / Deposition of new CENPA-containing nucleosomes at the centromere / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / RNA Polymerase I Promoter Opening / Inhibition of DNA recombination at telomere / negative regulation of receptor binding / Assembly of the ORC complex at the origin of replication / Meiotic synapsis / negative regulation of angiogenesis / early endosome lumen / DNA damage checkpoint signaling / Nef mediated downregulation of MHC class I complex cell surface expression / DAP12 interactions / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / positive regulation of DNA repair / DNA methylation / replication fork / Condensation of Prophase Chromosomes / transferrin transport / Chromatin modifications during the maternal to zygotic transition (MZT) / SIRT1 negatively regulates rRNA expression / condensed nuclear chromosome / cellular response to iron ion / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / PRC2 methylates histones and DNA / Endosomal/Vacuolar pathway / Regulation of endogenous retroelements by KRAB-ZFP proteins / male germ cell nucleus / Defective pyroptosis / meiotic cell cycle / Antigen Presentation: Folding, assembly and peptide loading of class I MHC / peptide antigen assembly with MHC class II protein complex / lumenal side of endoplasmic reticulum membrane / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / cellular response to iron(III) ion / RNA Polymerase I Promoter Escape / Nonhomologous End-Joining (NHEJ) / negative regulation of forebrain neuron differentiation / MHC class II protein complex / antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent / ER to Golgi transport vesicle membrane / peptide antigen assembly with MHC class I protein complex / regulation of iron ion transport / regulation of erythrocyte differentiation / Transcriptional regulation by small RNAs / HFE-transferrin receptor complex Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.5 Å MOLECULAR REPLACEMENT / Resolution: 2.5 Å | ||||||
 Authors Authors | Shiroishi, M. / Kuroki, K. / Rasubala, L. / Kohda, D. / Maenaka, K. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.Usa / Year: 2006 Journal: Proc.Natl.Acad.Sci.Usa / Year: 2006Title: Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d) Authors: Shiroishi, M. / Kuroki, K. / Rasubala, L. / Tsumoto, K. / Kumagai, I. / Kurimoto, E. / Kato, K. / Kohda, D. / Maenaka, K. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2dyp.cif.gz 2dyp.cif.gz | 128.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2dyp.ent.gz pdb2dyp.ent.gz | 99.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2dyp.json.gz 2dyp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2dyp_validation.pdf.gz 2dyp_validation.pdf.gz | 451.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2dyp_full_validation.pdf.gz 2dyp_full_validation.pdf.gz | 467.5 KB | Display | |
| Data in XML |  2dyp_validation.xml.gz 2dyp_validation.xml.gz | 24 KB | Display | |
| Data in CIF |  2dyp_validation.cif.gz 2dyp_validation.cif.gz | 32.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dy/2dyp https://data.pdbj.org/pub/pdb/validation_reports/dy/2dyp ftp://data.pdbj.org/pub/pdb/validation_reports/dy/2dyp ftp://data.pdbj.org/pub/pdb/validation_reports/dy/2dyp | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | The interaction between HLA-G (the molecule composed of chain A/B/C) and LILRB2 (chain D) is a biological unit. |
- Components
Components
| #1: Protein | Mass: 32017.492 Da / Num. of mol.: 1 / Fragment: residues in data base 25-300 / Mutation: C42S Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Plasmid: pGMT7 / Production host: Homo sapiens (human) / Plasmid: pGMT7 / Production host:  |
|---|---|
| #2: Protein | Mass: 11879.356 Da / Num. of mol.: 1 / Fragment: residues in data base 24-219 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Plasmid: pGMT7 / Production host: Homo sapiens (human) / Plasmid: pGMT7 / Production host:  |
| #3: Protein/peptide | Mass: 1148.424 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: This peptide was chemically synthesized / References: UniProt: P16104 |
| #4: Protein | Mass: 21896.688 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Plasmid: pGMT7 / Production host: Homo sapiens (human) / Plasmid: pGMT7 / Production host:  |
| #5: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.66 Å3/Da / Density % sol: 53.84 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 8 Details: 0.1M Tris-HCl pH8.0, 45% PEG400, VAPOR DIFFUSION, HANGING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SPring-8 SPring-8  / Beamline: BL41XU / Wavelength: 1 Å / Beamline: BL41XU / Wavelength: 1 Å |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Jun 20, 2005 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.5→50 Å / Num. obs: 25468 / % possible obs: 99.3 % / Observed criterion σ(I): 1 / Redundancy: 10.8 % / Biso Wilson estimate: 56.3 Å2 / Rmerge(I) obs: 0.052 / Net I/σ(I): 14.9 |
| Reflection shell | Resolution: 2.5→2.59 Å / Redundancy: 11 % / Rmerge(I) obs: 0.412 / Mean I/σ(I) obs: 3.5 / % possible all: 99.4 |
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2D31, 2GW5 Resolution: 2.5→46.66 Å / Rfactor Rfree error: 0.007 / Data cutoff high absF: 1719921.49 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / Stereochemistry target values: Maximum Likelihood
| ||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 44.1963 Å2 / ksol: 0.342353 e/Å3 | ||||||||||||||||||||
| Displacement parameters | Biso mean: 60.2 Å2
| ||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.5→46.66 Å
| ||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||
| LS refinement shell | Resolution: 2.5→2.66 Å / Rfactor Rfree error: 0.02 / Total num. of bins used: 6
|
 Movie
Movie Controller
Controller














 PDBj
PDBj















