[English] 日本語
 Yorodumi
Yorodumi- EMDB-3851: Near-atomic resolution fibril structure of complete amyloid-beta(... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3851 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Near-atomic resolution fibril structure of complete amyloid-beta(1-42) by cryo-EM | |||||||||
 Map data Map data | The density map was sharpened by a B-factor of -50 Ang^2 and filtered to 3.5 Ang. This density map was used for model building and refinement. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | amyloid / fibril / aggregation / Alzheimer's disease / Protein fibril | |||||||||
| Function / homology |  Function and homology information Function and homology informationamyloid-beta complex / growth cone lamellipodium / cellular response to norepinephrine stimulus / growth cone filopodium / microglia development / collateral sprouting in absence of injury / Formyl peptide receptors bind formyl peptides and many other ligands / axo-dendritic transport / regulation of Wnt signaling pathway / hippocampal neuron apoptotic process ...amyloid-beta complex / growth cone lamellipodium / cellular response to norepinephrine stimulus / growth cone filopodium / microglia development / collateral sprouting in absence of injury / Formyl peptide receptors bind formyl peptides and many other ligands / axo-dendritic transport / regulation of Wnt signaling pathway / hippocampal neuron apoptotic process / axon midline choice point recognition / astrocyte activation involved in immune response / regulation of synapse structure or activity / NMDA selective glutamate receptor signaling pathway / regulation of spontaneous synaptic transmission / mating behavior / growth factor receptor binding / peptidase activator activity / Golgi-associated vesicle / PTB domain binding / positive regulation of amyloid fibril formation / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / astrocyte projection / Lysosome Vesicle Biogenesis / neuron remodeling / Deregulated CDK5 triggers multiple neurodegenerative pathways in Alzheimer's disease models / nuclear envelope lumen / dendrite development / positive regulation of protein metabolic process / TRAF6 mediated NF-kB activation / negative regulation of long-term synaptic potentiation / signaling receptor activator activity / Advanced glycosylation endproduct receptor signaling / transition metal ion binding / main axon / The NLRP3 inflammasome / modulation of excitatory postsynaptic potential / regulation of multicellular organism growth / intracellular copper ion homeostasis / ECM proteoglycans / regulation of presynapse assembly / positive regulation of T cell migration / response to insulin-like growth factor stimulus / neuronal dense core vesicle / Purinergic signaling in leishmaniasis infection / positive regulation of chemokine production / Notch signaling pathway / cellular response to manganese ion / swimming behavior / clathrin-coated pit / extracellular matrix organization / neuron projection maintenance / astrocyte activation / positive regulation of calcium-mediated signaling / positive regulation of mitotic cell cycle / ionotropic glutamate receptor signaling pathway / Mitochondrial protein degradation / response to interleukin-1 / axonogenesis / protein serine/threonine kinase binding / platelet alpha granule lumen / regulation of neuron apoptotic process / cellular response to copper ion / cellular response to cAMP / positive regulation of glycolytic process / central nervous system development / dendritic shaft / endosome lumen / trans-Golgi network membrane / positive regulation of long-term synaptic potentiation / positive regulation of interleukin-1 beta production / learning / adult locomotory behavior / Post-translational protein phosphorylation / serine-type endopeptidase inhibitor activity / locomotory behavior / microglial cell activation / positive regulation of non-canonical NF-kappaB signal transduction / cellular response to nerve growth factor stimulus / TAK1-dependent IKK and NF-kappa-B activation / recycling endosome / synapse organization / regulation of long-term neuronal synaptic plasticity / visual learning / positive regulation of JNK cascade / Golgi lumen / response to lead ion / positive regulation of interleukin-6 production / cognition / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / cellular response to amyloid-beta / endocytosis / neuron projection development / positive regulation of tumor necrosis factor production / positive regulation of inflammatory response / calcium ion transport / Platelet degranulation / heparin binding / regulation of translation / regulation of gene expression Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
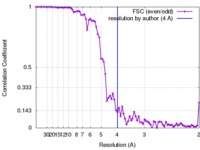
| Method | helical reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Gremer L / Schoelzel D | |||||||||
 Citation Citation |  Journal: Science / Year: 2017 Journal: Science / Year: 2017Title: Fibril structure of amyloid-β(1-42) by cryo-electron microscopy. Authors: Lothar Gremer / Daniel Schölzel / Carla Schenk / Elke Reinartz / Jörg Labahn / Raimond B G Ravelli / Markus Tusche / Carmen Lopez-Iglesias / Wolfgang Hoyer / Henrike Heise / Dieter ...Authors: Lothar Gremer / Daniel Schölzel / Carla Schenk / Elke Reinartz / Jörg Labahn / Raimond B G Ravelli / Markus Tusche / Carmen Lopez-Iglesias / Wolfgang Hoyer / Henrike Heise / Dieter Willbold / Gunnar F Schröder /   Abstract: Amyloids are implicated in neurodegenerative diseases. Fibrillar aggregates of the amyloid-β protein (Aβ) are the main component of the senile plaques found in brains of Alzheimer's disease ...Amyloids are implicated in neurodegenerative diseases. Fibrillar aggregates of the amyloid-β protein (Aβ) are the main component of the senile plaques found in brains of Alzheimer's disease patients. We present the structure of an Aβ(1-42) fibril composed of two intertwined protofilaments determined by cryo-electron microscopy (cryo-EM) to 4.0-angstrom resolution, complemented by solid-state nuclear magnetic resonance experiments. The backbone of all 42 residues and nearly all side chains are well resolved in the EM density map, including the entire N terminus, which is part of the cross-β structure resulting in an overall "LS"-shaped topology of individual subunits. The dimer interface protects the hydrophobic C termini from the solvent. The characteristic staggering of the nonplanar subunits results in markedly different fibril ends, termed "groove" and "ridge," leading to different binding pathways on both fibril ends, which has implications for fibril growth. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3851.map.gz emd_3851.map.gz | 36.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3851-v30.xml emd-3851-v30.xml emd-3851.xml emd-3851.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3851_fsc.xml emd_3851_fsc.xml | 7.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_3851.png emd_3851.png | 147.8 KB | ||
| Filedesc metadata |  emd-3851.cif.gz emd-3851.cif.gz | 5.5 KB | ||
| Others |  emd_3851_half_map_1.map.gz emd_3851_half_map_1.map.gz emd_3851_half_map_2.map.gz emd_3851_half_map_2.map.gz | 13.5 MB 13.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3851 http://ftp.pdbj.org/pub/emdb/structures/EMD-3851 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3851 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3851 | HTTPS FTP |
-Related structure data
| Related structure data |  5oqvMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3851.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3851.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The density map was sharpened by a B-factor of -50 Ang^2 and filtered to 3.5 Ang. This density map was used for model building and refinement. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.935 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: even half map.
| File | emd_3851_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | even half map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: odd half map.
| File | emd_3851_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | odd half map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Beta-amyloid protein 42 fibrils
| Entire | Name: Beta-amyloid protein 42 fibrils |
|---|---|
| Components |
|
-Supramolecule #1: Beta-amyloid protein 42 fibrils
| Supramolecule | Name: Beta-amyloid protein 42 fibrils / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Amyloid beta A4 protein
| Macromolecule | Name: Amyloid beta A4 protein / type: protein_or_peptide / ID: 1 / Number of copies: 9 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 4.520087 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DAEFRHDSGY EVHHQKLVFF AEDVGSNKGA IIGLMVGGVV IA UniProtKB: Amyloid-beta precursor protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 2 Component:
Details: in water | ||||||
|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil R 1.2/1.3 Quantifoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | ||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV Details: 2.5 microL sample was applied to the grid, blotted for 2.5 s before plunging.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 2026 / Average exposure time: 2.0 sec. / Average electron dose: 24.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 110000 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-5oqv: |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)