[English] 日本語
 Yorodumi
Yorodumi- EMDB-9734: Cryo-EM structure of the plant actin filaments from Zea mays pollen -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9734 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the plant actin filaments from Zea mays pollen | ||||||||||||||||||
 Map data Map data | None | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Microfilament / Helix / Actin / PROTEIN FIBRIL | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.9 Å | ||||||||||||||||||
 Authors Authors | Ren ZH / Zhang Y | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Plant Cell / Year: 2019 Journal: Plant Cell / Year: 2019Title: Cryo-EM Structure of Actin Filaments from Pollen. Authors: Zhanhong Ren / Yan Zhang / Yi Zhang / Yunqiu He / Pingzhou Du / Zhanxin Wang / Fei Sun / Haiyun Ren /  Abstract: Actins are among the most abundant and conserved proteins in eukaryotic cells, where they form filamentous structures that perform vital roles in key cellular processes. Although large amounts of ...Actins are among the most abundant and conserved proteins in eukaryotic cells, where they form filamentous structures that perform vital roles in key cellular processes. Although large amounts of data on the biochemical activities, dynamic behaviors, and important cellular functions of plant actin filaments have accumulated, their structural basis remains elusive. Here, we report a 3.9 Å structure of the plant actin filament from pollen (ZMPA) using cryo-electron microscopy. The structure shows a right-handed, double-stranded (two parallel strands) and staggered architecture that is stabilized by intra- and interstrand interactions. While the overall structure resembles that of other actin filaments, its DNase I binding loop bends farther outward, adopting an open conformation similar to that of the jasplakinolide- or beryllium fluoride (BeF)-stabilized rabbit skeletal muscle actin (RSMA) filament. Single-molecule magnetic tweezers analysis revealed that the ZMPA filament can resist a greater stretching force than the RSMA filament. Overall, these data provide evidence that plant actin filaments have greater stability than animal actin filaments, which might be important to their role as tracks for long-distance vesicle and organelle transportation.plantcell;31/12/2855/FX1F1fx1. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9734.map.gz emd_9734.map.gz | 27.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9734-v30.xml emd-9734-v30.xml emd-9734.xml emd-9734.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9734.png emd_9734.png | 134.5 KB | ||
| Filedesc metadata |  emd-9734.cif.gz emd-9734.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9734 http://ftp.pdbj.org/pub/emdb/structures/EMD-9734 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9734 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9734 | HTTPS FTP |
-Related structure data
| Related structure data |  6iugMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9734.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9734.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.063 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
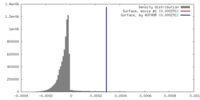
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The actin filament from Zea mays pollen
| Entire | Name: The actin filament from Zea mays pollen |
|---|---|
| Components |
|
-Supramolecule #1: The actin filament from Zea mays pollen
| Supramolecule | Name: The actin filament from Zea mays pollen / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Plant actin was purified from the pollen which was collected from maize (Zea mays) plants and polymerized at room temperature for 4 hours. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15 kDa/nm |
-Macromolecule #1: pollen F-actin
| Macromolecule | Name: pollen F-actin / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.211121 KDa |
| Sequence | String: IQPLVCDNGT GMVKAGFAGD DAPRAVFPSI VGRPRHTGVM VGMGQKDAYV GDEAQSKRGI LTLKYPIEHG IVSNWDDMEK IWHHTFYNE LRVAPEEHPV LLTEAPLNPK ANREKMTQIM FETFNTPAMY VAIQAVLSLY ASGRTTGIVL DSGDGVSHTV P IYEGYALP ...String: IQPLVCDNGT GMVKAGFAGD DAPRAVFPSI VGRPRHTGVM VGMGQKDAYV GDEAQSKRGI LTLKYPIEHG IVSNWDDMEK IWHHTFYNE LRVAPEEHPV LLTEAPLNPK ANREKMTQIM FETFNTPAMY VAIQAVLSLY ASGRTTGIVL DSGDGVSHTV P IYEGYALP HAILRLDLAG RDLTDYLMKI LTERGYSFTT TAEREIVRDM KEKLAYIALD YDQEMETAKT SSSVEKSYEL PD GQVITIG AERFRCPEVL FQPSFIGMEA AGIHETTYNS IMKCDVDIRK DLYGNIVLSG GTTMFPGIAD RMSKEITALA PSS MKIKVV APPERKYSVW IGGSILASLS TFQQMWIAKA EYDESGPSIV HRKCF UniProtKB: Actin-1 |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 5 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||||||||||||
| Grid | Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100.0 % / Chamber temperature: 298.0 K / Instrument: FEI VITROBOT MARK IV Details: Blotting time of 5.5 s and blotting force of level 2. | ||||||||||||||||||
| Details | Plant actin was dialyzed against buffer solution with pH 7.0 overnight in order to change the alkaline pH to the neutral pH and then polymerized at the final concentration of 0.4 mg/mL. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7676 pixel / Digitization - Dimensions - Height: 7420 pixel / Digitization - Frames/image: 2-27 / Number real images: 3605 / Average exposure time: 5.44 sec. / Average electron dose: 39.0 e/Å2 Details: A total of 3,605 micrographs were recorded at a calibrated pixel size of 1.063 angstrom. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.0 µm / Calibrated defocus min: 1.2 µm / Calibrated magnification: 22013 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: C / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Chain C from pdb 5OOC was rigid body fitted to plant actin map using Chimera; Then Phenix was used for real space refinement. Finally, filament with 5 real space refinement units were generated using helical parameter, and this filament with 5 units was real space refined using Phenix again. |
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT / Overall B value: 70 / Target criteria: Correlation coefficient |
| Output model |  PDB-6iug: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)