+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1kbc | ||||||
|---|---|---|---|---|---|---|---|
| Title | PROCARBOXYPEPTIDASE TERNARY COMPLEX | ||||||
 Components Components | NEUTROPHIL COLLAGENASE | ||||||
 Keywords Keywords | METALLOPROTEINASE / HYDROLYTIC ENZYME / COLLAGENASE / MATRIXIN / MMP-8 / HNC / INHIBITOR / HYDROLASE | ||||||
| Function / homology |  Function and homology information Function and homology informationneutrophil collagenase / tumor necrosis factor binding / positive regulation of microglial cell activation / positive regulation of neuroinflammatory response / positive regulation of tumor necrosis factor-mediated signaling pathway / Activation of Matrix Metalloproteinases / endodermal cell differentiation / Collagen degradation / collagen catabolic process / extracellular matrix disassembly ...neutrophil collagenase / tumor necrosis factor binding / positive regulation of microglial cell activation / positive regulation of neuroinflammatory response / positive regulation of tumor necrosis factor-mediated signaling pathway / Activation of Matrix Metalloproteinases / endodermal cell differentiation / Collagen degradation / collagen catabolic process / extracellular matrix disassembly / Degradation of the extracellular matrix / extracellular matrix organization / metalloendopeptidase activity / specific granule lumen / positive regulation of tumor necrosis factor production / tertiary granule lumen / peptidase activity / cellular response to lipopolysaccharide / collagen-containing extracellular matrix / endopeptidase activity / serine-type endopeptidase activity / Neutrophil degranulation / proteolysis / extracellular space / zinc ion binding / extracellular region Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.8 Å MOLECULAR REPLACEMENT / Resolution: 1.8 Å | ||||||
 Authors Authors | Betz, M. / Gomis-Rueth, F.X. / Bode, W. | ||||||
 Citation Citation |  Journal: Eur.J.Biochem. / Year: 1997 Journal: Eur.J.Biochem. / Year: 1997Title: 1.8-A crystal structure of the catalytic domain of human neutrophil collagenase (matrix metalloproteinase-8) complexed with a peptidomimetic hydroxamate primed-side inhibitor with a distinct selectivity profile. Authors: Betz, M. / Huxley, P. / Davies, S.J. / Mushtaq, Y. / Pieper, M. / Tschesche, H. / Bode, W. / Gomis-Ruth, F.X. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1kbc.cif.gz 1kbc.cif.gz | 87.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1kbc.ent.gz pdb1kbc.ent.gz | 65.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1kbc.json.gz 1kbc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1kbc_validation.pdf.gz 1kbc_validation.pdf.gz | 408.7 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1kbc_full_validation.pdf.gz 1kbc_full_validation.pdf.gz | 412.3 KB | Display | |
| Data in XML |  1kbc_validation.xml.gz 1kbc_validation.xml.gz | 9.4 KB | Display | |
| Data in CIF |  1kbc_validation.cif.gz 1kbc_validation.cif.gz | 14.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kb/1kbc https://data.pdbj.org/pub/pdb/validation_reports/kb/1kbc ftp://data.pdbj.org/pub/pdb/validation_reports/kb/1kbc ftp://data.pdbj.org/pub/pdb/validation_reports/kb/1kbc | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 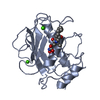
| ||||||||
| 2 | 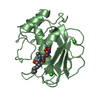
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 18258.918 Da / Num. of mol.: 2 / Fragment: CATALYTIC DOMAIN Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) Homo sapiens (human)Description: OBTAINED AS INCLUSION BODIES AND BY POSTERIOR REFOLDING Production host:  |
|---|
-Non-polymers , 5 types, 315 molecules 






| #2: Chemical | ChemComp-CA / #3: Chemical | ChemComp-ZN / #4: Chemical | #5: Chemical | #6: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.7 Å3/Da / Density % sol: 53.96 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Temperature: 22 ℃ / pH: 6 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction source | Wavelength: 1.5418 |
|---|---|
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.81→20 Å / Num. obs: 35353 / % possible obs: 96.6 % / Observed criterion σ(I): 3 / Redundancy: 4.3 % / Rmerge(I) obs: 0.066 |
| Reflection | *PLUS Num. measured all: 150360 |
| Reflection shell | *PLUS Highest resolution: 1.81 Å / Lowest resolution: 1.87 Å / % possible obs: 96.7 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.8→6 Å / MOLECULAR REPLACEMENT / Resolution: 1.8→6 Å /
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 25.7 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→6 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.1 / Classification: refinement X-PLOR / Version: 3.1 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller














 PDBj
PDBj








