-Search query
-Search result
Showing all 38 items for (author: zyla & ds)
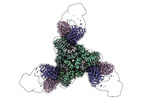
EMDB-42527: 
Pre-fusion Measles virus fusion protein complexed with Fab 77
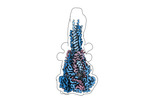
EMDB-42539: 
Structure of the Measles virus Fusion protein in the post-fusion conformation
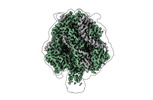
EMDB-42593: 
Structure of the Measles virus Fusion protein in the pre-fusion conformation

EMDB-42595: 
Structure of the Measles virus Fusion protein in the pre-fusion conformation with bound [FIP-HRC]2-PEG11
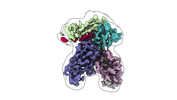
EMDB-43827: 
Fab 77-stabilized MeV F ectodomain fragment

EMDB-17863: 
2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod

EMDB-17878: 
2.5 A cryo-EM structure of the in vitro FimD-catalyzed assembly of type 1 pilus rod

EMDB-28756: 
Structure of SARS-CoV-2 Omicron BA.1 spike in complex with antibody Fab 1C3

EMDB-28757: 
Structure of SARS-CoV-2 spike with antibody Fabs 2A10 and 1H2 (Local refinement of the RBD and Fabs 1H2 and 2A10)

EMDB-28763: 
Negative stain EM map of SARS-CoV-2 D614G Spike in complex with 2A10 IgG

EMDB-28764: 
Negative stain EM map of SARS-CoV-2 D614G Spike in complex with 4H4 IgG

EMDB-28765: 
Negative stain EM map of SARS-CoV-2 D614G Spike in complex with 1C3 IgG

EMDB-28769: 
Negative stain EM map of SARS-CoV-2 Omicron BA.1 Spike in complex with 1C3 IgG

EMDB-28770: 
Negative stain EM map of SARS-CoV-2 D614G Spike in complex with 2G3 IgG

EMDB-28771: 
Negative stain EM map of SARS-CoV-2 D614G Spike in complex with 2E6 IgG

EMDB-28772: 
Negative stain EM map of SARS-CoV-2 D614G Spike in complex with 1H2 Fab

EMDB-28773: 
Negative stain EM map of SARS-CoV-2 D614G Spike in complex with 1G8 IgG

EMDB-27538: 
Lymphocytic choriomeningitis virus glycoprotein in complex with neutralizing antibody M28

EMDB-27539: 
Lymphocytic choriomeningitis virus glycoprotein

EMDB-10721: 
2.85 A cryo-EM structure of the in vivo assembled type 1 pilus rod

EMDB-11388: 
3.5 A cryo-EM structure of human uromodulin filament core

EMDB-11471: 
Extended cryo-EM map of native human uromodulin filament core at 4.7 A resolution

EMDB-11128: 
Cryo-electron tomogram of human Uromodulin filaments

EMDB-11129: 
Subtomogram average of human Uromodulin filaments

EMDB-11130: 
Cryo-electron tomogram of elastase treated human Uromodulin filaments (eUmod)

EMDB-11131: 
Subtomogram average of elastase treated human Uromodulin filaments (eUmod)

EMDB-11132: 
Cryo-electron tomogram of human Uromodulin filaments incubated with FimH lectin domain
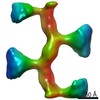
EMDB-11133: 
Subtomogram average of human Uromodulin filaments incubated with FimH lectin domain

EMDB-11134: 
Cryo-electron tomogram of elastase treated human Uromodulin filaments incubated with FimH lectin domain

EMDB-11135: 
Subtomogram average of elastase treated human Uromodulin fibers incubated with FimH lectin domain

EMDB-11136: 
Cryo-electron tomogram of E. coli AAEC [pSH2] incubated with human Uromodulin filaments

EMDB-11137: 
Cryo-electron tomogram of non-piliated E. coli AAEC189 incubated with human Uromodulin filaments

EMDB-11138: 
Cryo-electron tomogram of FIB milled E.coli - Uromodulin aggregates

EMDB-11139: 
Cryo-electron tomogram of E. coli AAEC189 [pSH2] incubated with elastase treated human Uromodulin filaments

EMDB-11140: 
Cryo-electron tomogram of urine from patient with E. coli urinary tract infection

EMDB-11141: 
Cryo-electron tomogram of urine from patient with K. pneumoniae urinary tract infection

EMDB-11142: 
Cryo-electron tomogram of urine from patient with P. aeruginosa urinary tract infection

EMDB-11143: 
Cryo-electron tomogram of urine from patient with S. mitis urinary tract infection
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
