+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
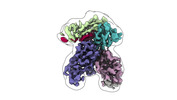
| Title | Fab 77-stabilized MeV F ectodomain fragment | ||||||||||||||||||
 Map data Map data | local filtered map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | VIRAL PROTEIN / glycoprotein / immune system / antibody / measles / high-resolution / antibody fragment / fab / neutralizing antibody / ectodomain | ||||||||||||||||||
| Function / homology | Precursor fusion glycoprotein F0, Paramyxoviridae / Fusion glycoprotein F0 / fusion of virus membrane with host plasma membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane / virion membrane / Fusion glycoprotein F0 Function and homology information Function and homology information | ||||||||||||||||||
| Biological species |   Measles virus strain Ichinose-B95a / Measles virus strain Ichinose-B95a /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | ||||||||||||||||||
 Authors Authors | Zyla D / Saphire EO | ||||||||||||||||||
| Funding support |  Switzerland, Switzerland,  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2024 Journal: Science / Year: 2024Title: A neutralizing antibody prevents postfusion transition of measles virus fusion protein. Authors: Dawid S Zyla / Roberta Della Marca / Gele Niemeyer / Gillian Zipursky / Kyle Stearns / Cameron Leedale / Elizabeth B Sobolik / Heather M Callaway / Chitra Hariharan / Weiwei Peng / Diptiben ...Authors: Dawid S Zyla / Roberta Della Marca / Gele Niemeyer / Gillian Zipursky / Kyle Stearns / Cameron Leedale / Elizabeth B Sobolik / Heather M Callaway / Chitra Hariharan / Weiwei Peng / Diptiben Parekh / Tara C Marcink / Ruben Diaz Avalos / Branka Horvat / Cyrille Mathieu / Joost Snijder / Alexander L Greninger / Kathryn M Hastie / Stefan Niewiesk / Anne Moscona / Matteo Porotto / Erica Ollmann Saphire /      Abstract: Measles virus (MeV) presents a public health threat that is escalating as vaccine coverage in the general population declines and as populations of immunocompromised individuals, who cannot be ...Measles virus (MeV) presents a public health threat that is escalating as vaccine coverage in the general population declines and as populations of immunocompromised individuals, who cannot be vaccinated, increase. There are no approved therapeutics for MeV. Neutralizing antibodies targeting viral fusion are one potential therapeutic approach but have not yet been structurally characterized or advanced to clinical use. We present cryo-electron microscopy (cryo-EM) structures of prefusion F alone [2.1-angstrom (Å) resolution], F complexed with a fusion-inhibitory peptide (2.3-Å resolution), F complexed with the neutralizing and protective monoclonal antibody (mAb) 77 (2.6-Å resolution), and an additional structure of postfusion F (2.7-Å resolution). In vitro assays and examination of additional EM classes show that mAb 77 binds prefusion F, arrests F in an intermediate state, and prevents transition to the postfusion conformation. These structures shed light on antibody-mediated neutralization that involves arrest of fusion proteins in an intermediate state. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43827.map.gz emd_43827.map.gz | 301.3 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43827-v30.xml emd-43827-v30.xml emd-43827.xml emd-43827.xml | 25.7 KB 25.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43827.png emd_43827.png | 47.1 KB | ||
| Masks |  emd_43827_msk_1.map emd_43827_msk_1.map | 15.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-43827.cif.gz emd-43827.cif.gz | 7.5 KB | ||
| Others |  emd_43827_additional_1.map.gz emd_43827_additional_1.map.gz emd_43827_half_map_1.map.gz emd_43827_half_map_1.map.gz emd_43827_half_map_2.map.gz emd_43827_half_map_2.map.gz | 14.7 MB 14.5 MB 14.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43827 http://ftp.pdbj.org/pub/emdb/structures/EMD-43827 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43827 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43827 | HTTPS FTP |
-Related structure data
| Related structure data |  9at8MC  8ut2C  8utfC  8uupC  8uuqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43827.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43827.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local filtered map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.403 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_43827_msk_1.map emd_43827_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
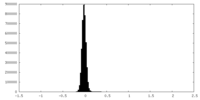
| Density Histograms |
-Additional map: sharpened map
| File | emd_43827_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
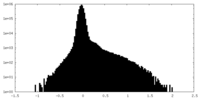
| Density Histograms |
-Half map: half-map 1
| File | emd_43827_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
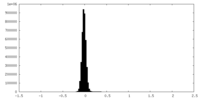
| Density Histograms |
-Half map: half-map 2
| File | emd_43827_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
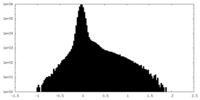
| Density Histograms |
- Sample components
Sample components
-Entire : Fab 77-stabilized MeV F ectodomain fragment
| Entire | Name: Fab 77-stabilized MeV F ectodomain fragment |
|---|---|
| Components |
|
-Supramolecule #1: Fab 77-stabilized MeV F ectodomain fragment
| Supramolecule | Name: Fab 77-stabilized MeV F ectodomain fragment / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: last refolding step of F ectodomain refolding in the presence of mAb 77 |
|---|---|
| Molecular weight | Theoretical: 43 KDa |
-Supramolecule #2: Antibody 77 Fab Heavy Chain, Antibody 77 Fab Light Chain
| Supramolecule | Name: Antibody 77 Fab Heavy Chain, Antibody 77 Fab Light Chain type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Measles virus Fusion glycoprotein
| Supramolecule | Name: Measles virus Fusion glycoprotein / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Measles virus strain Ichinose-B95a / Strain: Ichinose-B95a Measles virus strain Ichinose-B95a / Strain: Ichinose-B95a |
-Macromolecule #1: Fusion glycoprotein F0
| Macromolecule | Name: Fusion glycoprotein F0 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Measles virus strain Ichinose-B95a Measles virus strain Ichinose-B95a |
| Molecular weight | Theoretical: 57.379539 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGLKVNVSAI FMAVLLTLQT PTGQIHWGNL SKIGVVGIGS ASYKVMTRSS HQSLVIKLMP NITLLNNCTR VEIAEYRRLL RTVLEPIRD ALNAMTQNIR PVQSVASSRR HKRFAGVVLA GAALGVATAA QITAGIALHQ SMLNSQAIDN LRASLETTNQ A IEAIRQAG ...String: MGLKVNVSAI FMAVLLTLQT PTGQIHWGNL SKIGVVGIGS ASYKVMTRSS HQSLVIKLMP NITLLNNCTR VEIAEYRRLL RTVLEPIRD ALNAMTQNIR PVQSVASSRR HKRFAGVVLA GAALGVATAA QITAGIALHQ SMLNSQAIDN LRASLETTNQ A IEAIRQAG QGMILAVQGV QDYINNELIP SMNQLSCDLI GQKLGLKLLR YYTEILSLFG PSLRDPISAE ISIQALSYAL GG DINKVLE KLGYSGGDLL GILESRGIKA RITHVDTESY FIVLSIAYPT LSEIKGVIVH RLEGVSYNIG SQEWYTTVPK YVA TQGYLI SNFDESSCTF MPEGTVCSQN ALYPMSPLLQ ECLRGSTKSC ARTLVSGSFG NRFILSQGNL IANCASILCK CYTT GTIIN QDPDKILTYI AADHCPVVEV NGVTIQVGSR RYPDAVYLHR IDLGPPISLG RLDVGTNLGN AIAKLEDAKE LLESS DQIL RSMKGLSSTS IGVDDDDKAG WSHPQFEKGG GSGGGSGGGS WSHPQFEK UniProtKB: Fusion glycoprotein F0 |
-Macromolecule #2: mAb 77 light chain
| Macromolecule | Name: mAb 77 light chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.532512 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGWSCIILFL VATATGVHSD VQITQSPSYL AASPGETITI NCRASKSISK YLAWYQEKPG KTNELLIYSG STLQSGIPSR FRGSGSGTD FTLTISSLEP EDFAMYYCQQ HNEYTLTFGG GTKLELKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY P REAKVQWK ...String: MGWSCIILFL VATATGVHSD VQITQSPSYL AASPGETITI NCRASKSISK YLAWYQEKPG KTNELLIYSG STLQSGIPSR FRGSGSGTD FTLTISSLEP EDFAMYYCQQ HNEYTLTFGG GTKLELKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY P REAKVQWK VDNALQSGNS QESVTEQDSK DSTYSLSSTL TLSKADYEKH KVYACEVTHQ GLSSPVTKSF NRGEC |
-Macromolecule #3: mAB 77 heavy chain
| Macromolecule | Name: mAB 77 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.722336 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGWSCIILFL VATATGVHSD VQLQESGPGL VKPSQSLSLT CTVSGYSITS DYAWNWIRQF PGNKLEWMGY ISYTLTTGYN PSLKSRISI TRDSSKNQFF LQLNSVTTED TATYYCARSG WLLPYWYFDV WGAGTTVTVS SASTKGPSVF PLAPSSKSTS G GTAALGCL ...String: MGWSCIILFL VATATGVHSD VQLQESGPGL VKPSQSLSLT CTVSGYSITS DYAWNWIRQF PGNKLEWMGY ISYTLTTGYN PSLKSRISI TRDSSKNQFF LQLNSVTTED TATYYCARSG WLLPYWYFDV WGAGTTVTVS SASTKGPSVF PLAPSSKSTS G GTAALGCL VKDYFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV EP KSCDKGL EVLFQGPTHT CPPCPAPELL GGPSVFLFPP KPKDTLMISR TPEVTCVVVD VSHEDPEVKF NWYVDGVEVH NAK TKPREE QYNSTYRVVS VLTVLHQDWL NGKEYKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS RDELTKNQVS LTCL VKGFY PSDIAVEWES NGQPENNYKT TPPVLDSDGS FFLYSKLTVD KSRWQQGNVF SCSVMHEALH NHYTQKSLSL SPGK |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: HEPES 50 mM, pH 8.0, NaCl 150 mM |
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 4 / Number real images: 10344 / Average exposure time: 2.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)