[English] 日本語
 Yorodumi
Yorodumi- EMDB-28756: Structure of SARS-CoV-2 Omicron BA.1 spike in complex with antibo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
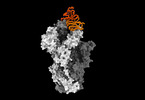
| Title | Structure of SARS-CoV-2 Omicron BA.1 spike in complex with antibody Fab 1C3 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VIRAL PROTEIN / glycoprotein / immune system / antibody / SARS-CoV-2 / COVID / VIRAL PROTEIN-Immune System complex / coronavirus | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.35 Å | |||||||||
 Authors Authors | Yu X / Zyla D / Hastie KM / Saphire EO | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2023 Journal: Cell Rep / Year: 2023Title: Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence. Authors: Kathryn M Hastie / Xiaoying Yu / Fernanda Ana-Sosa-Batiz / Dawid S Zyla / Stephanie S Harkins / Chitra Hariharan / Hal Wasserman / Michelle A Zandonatti / Robyn Miller / Erin Maule / Kenneth ...Authors: Kathryn M Hastie / Xiaoying Yu / Fernanda Ana-Sosa-Batiz / Dawid S Zyla / Stephanie S Harkins / Chitra Hariharan / Hal Wasserman / Michelle A Zandonatti / Robyn Miller / Erin Maule / Kenneth Kim / Kristen M Valentine / Sujan Shresta / Erica Ollmann Saphire /  Abstract: Therapeutic antibodies are an important tool in the arsenal against coronavirus infection. However, most antibodies developed early in the pandemic have lost most or all efficacy against newly ...Therapeutic antibodies are an important tool in the arsenal against coronavirus infection. However, most antibodies developed early in the pandemic have lost most or all efficacy against newly emergent strains of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), particularly those of the Omicron lineage. Here, we report the identification of a panel of vaccinee-derived antibodies that have broad-spectrum neutralization activity. Structural and biochemical characterization of the three broadest-spectrum antibodies reveal complementary footprints and differing requirements for avidity to overcome variant-associated mutations in their binding footprints. In the K18 mouse model of infection, these three antibodies exhibit protective efficacy against BA.1 and BA.2 infection. This study highlights the resilience and vulnerabilities of SARS-CoV-2 antibodies and provides road maps for further development of broad-spectrum therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28756.map.gz emd_28756.map.gz | 123.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28756-v30.xml emd-28756-v30.xml emd-28756.xml emd-28756.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28756_fsc.xml emd_28756_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_28756.png emd_28756.png | 41 KB | ||
| Filedesc metadata |  emd-28756.cif.gz emd-28756.cif.gz | 6.8 KB | ||
| Others |  emd_28756_half_map_1.map.gz emd_28756_half_map_1.map.gz emd_28756_half_map_2.map.gz emd_28756_half_map_2.map.gz | 226.4 MB 226.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28756 http://ftp.pdbj.org/pub/emdb/structures/EMD-28756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28756 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28756 | HTTPS FTP |
-Related structure data
| Related structure data |  8f0gMC  8e1gC  8f0hC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28756.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28756.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||
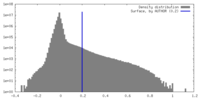
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28756_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
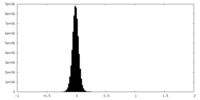
| Density Histograms |
-Half map: #1
| File | emd_28756_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
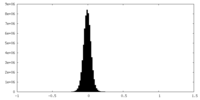
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of SARS-CoV-2 Omicron BA.1 spike in complex with antibo...
| Entire | Name: Structure of SARS-CoV-2 Omicron BA.1 spike in complex with antibody 1C3 Fab |
|---|---|
| Components |
|
-Supramolecule #1: Structure of SARS-CoV-2 Omicron BA.1 spike in complex with antibo...
| Supramolecule | Name: Structure of SARS-CoV-2 Omicron BA.1 spike in complex with antibody 1C3 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: Antibody 1C3 Fab Heavy Chain,Antibody 1C3 Fab Light Chain
| Supramolecule | Name: Antibody 1C3 Fab Heavy Chain,Antibody 1C3 Fab Light Chain type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Spike glycoprotein
| Supramolecule | Name: Spike glycoprotein / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Antibody 1C3 Fab Heavy Chain
| Macromolecule | Name: Antibody 1C3 Fab Heavy Chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.718235 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLLESGGG LVQPGRSLRL SCAASGFTFD DYAMHWVRQP PGKGLEWVSG SSWNSGSVVY ADSVKGRFTI SRDSAKNSLH LQMNSLRVE DTALYYCAKA VDPTRGSYSP DYGFDIWGQG TMVTVSS |
-Macromolecule #2: Antibody 1C3 Fab Light Chain
| Macromolecule | Name: Antibody 1C3 Fab Light Chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.554764 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DAIRMTQSPS SLSASVGDRV TITCRASQSI SSYLNWYQQK PGKAPNLLIY AASSLESGVP SRFSGSGSGT DFTLTISSLQ PEDFATYYC QQSYSTPLTF GGGTKVEIK |
-Macromolecule #3: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 134.224359 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGVKVLFALI CIAVAEAQCV NLTTRTQLPP AYTNSFTRGV YYPDKVFRSS VLHSTQDLFL PFFSNVTWFH VISGTNGTKR FDNPVLPFN DGVYFASIEK SNIIRGWIFG TTLDSKTQSL LIVNNATNVV IKVCEFQFCN DPFLDHKNNK SWMESEFRVY S SANNCTFE ...String: MGVKVLFALI CIAVAEAQCV NLTTRTQLPP AYTNSFTRGV YYPDKVFRSS VLHSTQDLFL PFFSNVTWFH VISGTNGTKR FDNPVLPFN DGVYFASIEK SNIIRGWIFG TTLDSKTQSL LIVNNATNVV IKVCEFQFCN DPFLDHKNNK SWMESEFRVY S SANNCTFE YVSQPFLMDL EGKQGNFKNL REFVFKNIDG YFKIYSKHTP IIVREPEDLP QGFSALEPLV DLPIGINITR FQ TLLALHR SYLTPGDSSS GWTAGAAAYY VGYLQPRTFL LKYNENGTIT DAVDCALDPL SETKCTLKSF TVEKGIYQTS NFR VQPTES IVRFPNITNL CPFDEVFNAT RFASVYAWNR KRISNCVADY SVLYNLAPFF TFKCYGVSPT KLNDLCFTNV YADS FVIRG DEVRQIAPGQ TGNIADYNYK LPDDFTGCVI AWNSNKLDSK VSGNYNYLYR LFRKSNLKPF ERDISTEIYQ AGNKP CNGV AGFNCYFPLR SYSFRPTYGV GHQPYRVVVL SFELLHAPAT VCGPKKSTNL VKNKCVNFNF NGLKGTGVLT ESNKKF LPF QQFGRDIADT TDAVRDPQTL EILDITPCSF GGVSVITPGT NTSNQVAVLY QGVNCTEVPV AIHADQLTPT WRVYSTG SN VFQTRAGCLI GAEYVNNSYE CDIPIGAGIC ASYQTQTKSH GSASSVASQS IIAYTMSLGA ENSVAYSNNS IAIPTNFT I SVTTEILPVS MTKTSVDCTM YICGDSTECS NLLLQYGSFC TQLKRALTGI AVEQDKNTQE VFAQVKQIYK TPPIKYFGG FNFSQILPDP SKPSKRSPIE DLLFNKVTLA DAGFIKQYGD CLGDIAARDL ICAQKFKGLT VLPPLLTDEM IAQYTSALLA GTITSGWTF GAGPALQIPF PMQMAYRFNG IGVTQNVLYE NQKLIANQFN SAIGKIQDSL SSTPSALGKL QDVVNHNAQA L NTLVKQLS SKFGAISSVL NDIFSRLDPP EAEVQIDRLI TGRLQSLQTY VTQQLIRAAE IRASANLAAT KMSECVLGQS KR VDFCGKG YHLMSFPQSA PHGVVFLHVT YVPAQEKNFT TAPAICHDGK AHFPREGVFV SNGTHWFVTQ RNFYEPQIIT TDN TFVSGN CDVVIGIVNN TVYDPLQPEL DSFKEELDKY FKNHTSPDVD LGDISGINAS VVNIQKEIDR LNEVAKNLNE SLID LQELG KYEQ UniProtKB: Spike glycoprotein |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 18 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: TBS buffer pH 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR Details: home-made GO grids with continues graphene oxide layer |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)