-Search query
-Search result
Showing 1 - 50 of 343 items for (author: zimmer & g)
EMDB-18036: 
In situ structure of E. coli 70S ribosome
EMDB-18037: 
In situ 70S ribosome of E. coli K-12 untreated cells

EMDB-18038: 
In situ 70S ribosome of E. coli K-12 cells treated with tetracycline

EMDB-18039: 
In situ 70S ribosome of E. coli ED1a untreated cells

EMDB-18040: 
In situ 70S ribosome of E. coli ED1a cells treated with tetracycline
EMDB-18041: 
E. coli K-12 70S ribosome bound to mRNA A-tRNA, P-tRNA, E-tRNA
EMDB-18042: 
E. coli ED1a 70S ribosome bound to mRNA A-tRNA, P-tRNA, E-tRNA

EMDB-19206: 
E. coli ED1a 70S-tetracycline complex - focused refinement on 30S head

EMDB-19207: 
E. coli ED1a 70S-tetracycline complex - focused refinement on 30S body

EMDB-19208: 
E. coli ED1a 70S-tetracycline complex - focused refinement on 50S
EMDB-40622: 
Chlorella virus Hyaluronan Synthase bound to GlcA extended GlcNAc primer

EMDB-40623: 
Chlorella virus Hyaluronan Synthase bound to GlcNAc primer and UDP-GlcA

EMDB-40624: 
Chlorella virus Hyaluronan Synthase bound to GlcA extended GlcNAc primer and UDP

EMDB-40591: 
Xenopus laevis hyaluronan synthase 1

EMDB-40594: 
Xenopus laevis hyaluronan synthase 1, nascent HA polymer bound state
EMDB-40598: 
Xenopus laevis hyaluronan synthase 1, UDP-bound, gating loop inserted state

PDB-8smm: 
Xenopus laevis hyaluronan synthase 1

PDB-8smn: 
Xenopus laevis hyaluronan synthase 1, nascent HA polymer bound state

PDB-8smp: 
Xenopus laevis hyaluronan synthase 1, UDP-bound, gating loop inserted state

EMDB-40477: 
KLHDC2 in complex with EloB and EloC

PDB-8sh2: 
KLHDC2 in complex with EloB and EloC

EMDB-15964: 
Subtomogram averaging of SARS-CoV-2 nsp3-4 delta Ubl1-Mac1 obtained from cryo-ET of cryo-FIB milled VeroE6 cells transfected with nsp3-4 delta Ubl1-Mac1

EMDB-15965: 
Subtomogram average of SARS-CoV-2 nsp3-4 delate Ubl1-Ubl2 obtained from cryo-ET of cryo-FIB milled VeroE6 cells transfected with nsp3-4 delta Ubl1-Ubl2

EMDB-40711: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor

PDB-8sqn: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor

EMDB-27272: 
CryoEM structure of Western equine encephalitis virus VLP

PDB-8daq: 
CryoEM structure of Western equine encephalitis virus VLP
EMDB-27271: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor

PDB-8dan: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor

EMDB-17208: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN

EMDB-17212: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant

EMDB-17249: 
CRYO-EM CONSENSUS MAP OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
EMDB-17250: 
CRYO-EM FOCUSED REFINEMENT MAPS OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN

EMDB-17254: 
CRYO-EM CONSENSUS MAP OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 SKO
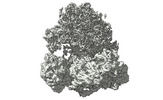
EMDB-17255: 
CRYO-EM FOCUSED REFINEMENT OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant

PDB-8ova: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN

PDB-8ove: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant

EMDB-15963: 
Subtomogram average SARS-CoV-2 nsp3-4 from cryo-ET of VeroE6 expressing nsp3-4 (filtered to 20A resolution).

EMDB-28644: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor

PDB-8ewf: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor

EMDB-15925: 
Cryo-electron tomogram acquired on cryo-lamella of VeroE6 cells transfected with SARS-CoV-2 HA-nsp3-4-V5, plunge frozen at 16 hpt

EMDB-15926: 
Cryo-electron tomogram of VeroE6 cells transfected with HA-nsp3-deltaUbl1-Ubl2-nsp4-V5, plunge frozen at 16 hpt

EMDB-15927: 
Cryo-electron tomogram of VeroE6 cells transfected with HA-nsp3-deltaUbl1-Mac1-nsp4-V5. Cells were plunge-frozen at 16h post-transfection and subjected to cryo-FIB milling.

EMDB-15928: 
Cryo-electron tomogram of VeroE6 cells transfected with HA-GG>AA-nsp4-V5, plunge-frozen at 16 hpt and subjected to cryo-FIB milling.

EMDB-15929: 
Cryo-electron tomogram of VeroE6 cells transfected with nsp3-4, vitrified by plunge-freezing at 16 hours post transfection and processed by cryo-FIB milling.

EMDB-15264: 
IF(apo/as isolated) conformation of CydDC mutant (H85A.C) in AMP-PNP(CydD) bound state (Dataset-17)
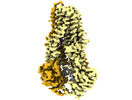
EMDB-15265: 
IF(heme/coordinated) conformation of CydDC mutant (E500Q.C) in AMP-PNP(CydC)/AMP-PNP(CydD) bound state (Dataset-23)

EMDB-16304: 
In situ outer dynein arm from Chlamydomonas reinhardtii in a pre-power stroke state

EMDB-16312: 
In situ outer dynein arm from Chlamydomonas reinhardtii in the post-power stroke state

PDB-8bwy: 
In situ outer dynein arm from Chlamydomonas reinhardtii in a pre-power stroke state
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
