-Search query
-Search result
Showing 1 - 50 of 243 items for (author: zhang & rz)

EMDB-35192: 
A cryoEM structure of the dimer of (S)-carbonyl reductase II

EMDB-29442: 
Subtomogram of the 4-beta-ring hoop of M. hungatei sheath structure.

EMDB-29443: 
Subtomogram average of 3-beta-ring hoop structure of M. hungatei sheath

EMDB-29448: 
Subtomogram average of 5-beta-ring hoop structure of M. hungatei sheath

EMDB-16603: 
Type2 alpha-synuclein filament assembled in vitro by wild-type and mutant (7 residues insertion) protein

EMDB-16604: 
Alpha-synuclein filament assembled in vitro with mutant (7 residues insertion) protein

EMDB-16608: 
WT alpha-synuclein filament assembled in vitro

PDB-8ceb: 
Type2 alpha-synuclein filament assembled in vitro by wild-type and mutant (7 residues insertion) protein

EMDB-16600: 
Type1 alpha-synuclein filament assembled in vitro by wild-type and mutant (7 residues insertion) protein

PDB-8ce7: 
Type1 alpha-synuclein filament assembled in vitro by wild-type and mutant (7 residues insertion) protein

EMDB-16188: 
Cryo-EM structure of alpha-synuclein singlet filament from Juvenile-onset synucleinopathy

EMDB-16189: 
Cryo-EM structure of alpha-synuclein filaments doublet from Juvenile-onset synucleinopathy

PDB-8bqv: 
Cryo-EM structure of alpha-synuclein singlet filament from Juvenile-onset synucleinopathy

PDB-8bqw: 
Cryo-EM structure of alpha-synuclein filaments doublet from Juvenile-onset synucleinopathy

EMDB-16022: 
Amyloid-beta 42 filaments extracted from the human brain with Arctic mutation (E22G) of Alzheimer's disease | ABeta42

EMDB-16023: 
Amyloid-beta tetrameric filaments with the Arctic mutation (E22G) from Alzheimer's disease brains | ABeta40

EMDB-16027: 
Murine amyloid-beta filaments with the Arctic mutation (E22G) from APP(NL-G-F) mouse brains | ABeta

PDB-8bfz: 
Amyloid-beta 42 filaments extracted from the human brain with Arctic mutation (E22G) of Alzheimer's disease | ABeta42

PDB-8bg0: 
Amyloid-beta tetrameric filaments with the Arctic mutation (E22G) from Alzheimer's disease brains | ABeta40

PDB-8bg9: 
Murine amyloid-beta filaments with the Arctic mutation (E22G) from APP(NL-G-F) mouse brains | ABeta

EMDB-16434: 
Type Ib beta-amyloid 42 Filaments from Human Brain

EMDB-26401: 
SARS-CoV-2 spike trimer in complex with Fab NE12, ensemble map

EMDB-26402: 
SARS-CoV-2 spike trimer in complex with Fab NE12, local refinement map

EMDB-26403: 
SARS-CoV-2 spike trimer in complex with Fab NA8, ensemble map

EMDB-26404: 
SARS-CoV-2 spike trimer in complex with Fab NA8, local refinement map

PDB-7u9o: 
SARS-CoV-2 spike trimer RBD in complex with Fab NE12

PDB-7u9p: 
SARS-CoV-2 spike trimer RBD in complex with Fab NA8

EMDB-27730: 
SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to linker variant of affinity matured ACE2 mimetic CVD432

EMDB-27731: 
SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to computationally engineered ACE2 mimetic CVD293

PDB-8dv1: 
SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to linker variant of affinity matured ACE2 mimetic CVD432

PDB-8dv2: 
SARS-CoV-2 Wuhan-hu-1-Spike-RBD bound to computationally engineered ACE2 mimetic CVD293

EMDB-15285: 
Cryo-EM structure of alpha-synuclein filaments from Parkinson's disease and dementia with Lewy bodies

PDB-8a9l: 
Cryo-EM structure of alpha-synuclein filaments from Parkinson's disease and dementia with Lewy bodies

EMDB-33748: 
Spike_GSAS_6P protomer RBD domain bound with R1-32 Fab and ACE2 with 3:3:3 ratio
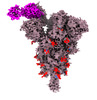
EMDB-33760: 
Spike_GSAS_6P and R1-32 Fab with 3to1 ratio
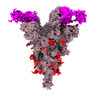
EMDB-33764: 
SARS-COV-2 Spike_GSAS_6P bound with two R1-32 Fabs
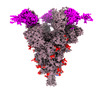
EMDB-33766: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs

EMDB-33772: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2

PDB-7ydi: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region

PDB-7ydy: 
SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab

PDB-7ye5: 
SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs

PDB-7ye9: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs

PDB-7yeg: 
SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2

EMDB-33453: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation

EMDB-33454: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-211 Conformation

EMDB-33455: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation

EMDB-33456: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation

EMDB-33457: 
Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation

EMDB-33458: 
Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation

EMDB-33459: 
Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
