-Search query
-Search result
Showing 1 - 50 of 10,397 items for (author: yu & b)
EMDB-42603: 
Human p97/VCP structure with a triazole inhibitor (NSC799462/hexamer)
EMDB-42625: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC804515)
EMDB-42626: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC819701/up)
EMDB-42627: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC819701/down)
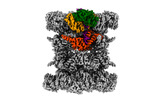
EMDB-44748: 
Human p97/VCP structure with a triazole inhibitor (NSC799462/dodecamer)

EMDB-41766: 
CryoEM structure of D2 dopamine receptor in complex with GoA KE mutant, scFv16, and dopamine

EMDB-41776: 
CryoEM structure of D2 dopamine receptor in complex with GoA KE mutant and dopamine

EMDB-44551: 
Map of eastern equine encephalitis virus q3 spike protein in complex with VLDLR without masked refinement
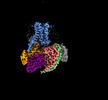
EMDB-60628: 
Carazolol-activated human beta3 adrenergic receptor
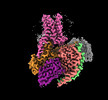
EMDB-60629: 
Epinephrine-activated human beta3 adrenergic receptor

EMDB-31367: 
Structure of mumps virus nucleoprotein without C-arm

EMDB-50124: 
Mammalian ternary complex of a translating 80S ribosome, NAC and NatA/E

EMDB-50125: 
Mammalian quaternary complex of a translating 80S ribosome, NAC, MetAP1 and NatA/E

EMDB-50126: 
Mammalian quaternary complex of a translating 80S ribosome, NAC, MetAP1 and NatA/E-HYPK

EMDB-38850: 
Cryo-EM structure of human dopamine transporter in apo state

EMDB-38851: 
Cryo-EM structure of human dopamine transporter in complex with dopamine

EMDB-38852: 
Cryo-EM structure of human dopamine transporter in complex with benztropine

EMDB-38853: 
structure of a proteinACryo-EM structure of human dopamine transporter in complex with GBR12909

EMDB-38854: 
Cryo-EM structure of human dopamine transporter in complex with methylphenidate

EMDB-42050: 
Structure of eastern equine encephalitis virus VLP in complex with VLDLR LA1

EMDB-42055: 
Structure of eastern equine encephalitis virus VLP unliganded quasi-threefold spike protein

EMDB-38845: 
Icosahedrally averaged cryo-EM reconstruction of PhiKZ capsid before applying the "block-based" reconstruction method

EMDB-38846: 
Block 1 of PhiKZ capsid

EMDB-38848: 
Block 2 of PhiKZ capsid

EMDB-39002: 
Composite cryo-EM map of PhiKZ capsid after applying the "block-based" reconstruction method

PDB-8y6v: 
Near-atomic structure of icosahedrally averaged jumbo bacteriophage PhiKZ capsid

EMDB-39212: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH8.0 (3.23A)

EMDB-39213: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH6.5 (2.82A)

EMDB-39214: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH5.0 (3.52A)

EMDB-39215: 
Cryo-EM structure of Dragon Grouper nervous necrosis virion at pH6.5 (3.12A)

EMDB-39217: 
Cryo-EM structure of Dragon Grouper nervous necrosis virion at pH5.0 (4.36A)

PDB-8yf6: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH8.0 (3.23A)

PDB-8yf7: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH6.5 (2.82A)

PDB-8yf8: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH5.0 (3.52A)

PDB-8yf9: 
Cryo-EM structure of Dragon Grouper nervous necrosis virion at pH6.5 (3.12A)

EMDB-43814: 
Cryo-EM structure of the active Lactococcus lactis Csm bound to target in post-cleavage stage

EMDB-43815: 
Cryo-EM structure of the active Lactococcus lactis Csm bound to target in pre-cleavage stage

PDB-9ash: 
Cryo-EM structure of the active Lactococcus lactis Csm bound to target in post-cleavage stage

PDB-9asi: 
Cryo-EM structure of the active Lactococcus lactis Csm bound to target in pre-cleavage stage

EMDB-45005: 
Paired Helical Filament of tau amyloids found in Down Syndrome individuals

EMDB-45007: 
Straight Filament of tau amyloids found in Down Syndrome individuals

EMDB-45008: 
Paired Helical Filaments purified from Down Syndrome individual brain tissue applied to graphene oxide antibody affinity grids

EMDB-45009: 
Straight Filaments purified from Down Syndrome individual brain tissue applied to graphene oxide antibody affinity grids
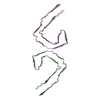
PDB-9bxi: 
Paired Helical Filament of tau amyloids found in Down Syndrome individuals
PDB-9bxo: 
Straight Filament of tau amyloids found in Down Syndrome individuals
PDB-9bxq: 
Paired Helical Filaments purified from Down Syndrome individual brain tissue applied to graphene oxide antibody affinity grids

PDB-9bxr: 
Straight Filaments purified from Down Syndrome individual brain tissue applied to graphene oxide antibody affinity grids
EMDB-44246: 
Cryo-EM structure of HIV-1 JRFL v6 Env in complex with vaccine-elicited, Membrane Proximal External Region (MPER) directed antibody DH1317.4.

EMDB-41854: 
Structure of Human Mitochondrial Chaperonin V72I Mutant

PDB-8u39: 
Structure of Human Mitochondrial Chaperonin V72I mutant
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
