[English] 日本語
 Yorodumi
Yorodumi- EMDB-41766: CryoEM structure of D2 dopamine receptor in complex with GoA KE m... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of D2 dopamine receptor in complex with GoA KE mutant, scFv16, and dopamine | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / Dopamine / DRD2 / Dominant Negative / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of dopamine receptor signaling pathway / positive regulation of dopamine uptake involved in synaptic transmission / negative regulation of dephosphorylation / positive regulation of glial cell-derived neurotrophic factor production / acid secretion / dopamine neurotransmitter receptor activity, coupled via Gi/Go / nervous system process involved in regulation of systemic arterial blood pressure / response to histamine / negative regulation of circadian sleep/wake cycle, sleep / regulation of synapse structural plasticity ...negative regulation of dopamine receptor signaling pathway / positive regulation of dopamine uptake involved in synaptic transmission / negative regulation of dephosphorylation / positive regulation of glial cell-derived neurotrophic factor production / acid secretion / dopamine neurotransmitter receptor activity, coupled via Gi/Go / nervous system process involved in regulation of systemic arterial blood pressure / response to histamine / negative regulation of circadian sleep/wake cycle, sleep / regulation of synapse structural plasticity / regulation of locomotion involved in locomotory behavior / neuron-neuron synaptic transmission / negative regulation of dopamine secretion / adenohypophysis development / negative regulation of cellular response to hypoxia / hyaloid vascular plexus regression / adenylate cyclase-inhibiting dopamine receptor signaling pathway / cerebral cortex GABAergic interneuron migration / response to inactivity / orbitofrontal cortex development / regulation of potassium ion transport / Dopamine receptors / negative regulation of neuron migration / dopamine binding / branching morphogenesis of a nerve / regulation of dopamine uptake involved in synaptic transmission / phospholipase C-activating dopamine receptor signaling pathway / positive regulation of growth hormone secretion / peristalsis / heterotrimeric G-protein binding / drinking behavior / G protein-coupled receptor complex / grooming behavior / behavioral response to ethanol / positive regulation of renal sodium excretion / auditory behavior / striatum development / positive regulation of G protein-coupled receptor signaling pathway / dopaminergic synapse / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / positive regulation of multicellular organism growth / G protein-coupled receptor internalization / non-motile cilium / heterocyclic compound binding / vesicle docking involved in exocytosis / response to iron ion / G protein-coupled dopamine receptor signaling pathway / negative regulation of synaptic transmission, glutamatergic / positive regulation of urine volume / adult walking behavior / arachidonate secretion / ciliary membrane / negative regulation of cytosolic calcium ion concentration / positive regulation of neuroblast proliferation / response to morphine / regulation of synaptic transmission, GABAergic / temperature homeostasis / positive regulation of cytokinesis / regulation of heart contraction / pigmentation / dopamine metabolic process / dopamine uptake involved in synaptic transmission / parallel fiber to Purkinje cell synapse / regulation of dopamine secretion / positive regulation of receptor internalization / cellular response to ethanol / response to light stimulus / lateral plasma membrane / associative learning / neuroblast proliferation / G-protein alpha-subunit binding / negative regulation of protein secretion / endocytic vesicle / prepulse inhibition / potassium channel regulator activity / long-term memory / sperm flagellum / response to axon injury / postsynaptic modulation of chemical synaptic transmission / negative regulation of blood pressure / regulation of sodium ion transport / behavioral response to cocaine / cellular response to retinoic acid / synapse assembly / release of sequestered calcium ion into cytosol / ionotropic glutamate receptor binding / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / presynaptic modulation of chemical synaptic transmission / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / axon terminus / acrosomal vesicle / axonogenesis / regulation of heart rate / negative regulation of innate immune response / muscle contraction / negative regulation of cell migration / epithelial cell proliferation Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Krumm BE / Kapolka NJ / Fay JF / Roth BL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: A neurodevelopmental disorder mutation locks G proteins in the transitory pre-activated state. Authors: Kevin M Knight / Brian E Krumm / Nicholas J Kapolka / W Grant Ludlam / Meng Cui / Sepehr Mani / Iya Prytkova / Elizabeth G Obarow / Tyler J Lefevre / Wenyuan Wei / Ning Ma / Xi-Ping Huang / ...Authors: Kevin M Knight / Brian E Krumm / Nicholas J Kapolka / W Grant Ludlam / Meng Cui / Sepehr Mani / Iya Prytkova / Elizabeth G Obarow / Tyler J Lefevre / Wenyuan Wei / Ning Ma / Xi-Ping Huang / Jonathan F Fay / Nagarajan Vaidehi / Alan V Smrcka / Paul A Slesinger / Diomedes E Logothetis / Kirill A Martemyanov / Bryan L Roth / Henrik G Dohlman /  Abstract: Many neurotransmitter receptors activate G proteins through exchange of GDP for GTP. The intermediate nucleotide-free state has eluded characterization, due largely to its inherent instability. Here ...Many neurotransmitter receptors activate G proteins through exchange of GDP for GTP. The intermediate nucleotide-free state has eluded characterization, due largely to its inherent instability. Here we characterize a G protein variant associated with a rare neurological disorder in humans. Gα has a charge reversal that clashes with the phosphate groups of GDP and GTP. As anticipated, the purified protein binds poorly to guanine nucleotides yet retains wild-type affinity for G protein βγ subunits. In cells with physiological concentrations of nucleotide, Gα forms a stable complex with receptors and Gβγ, impeding effector activation. Further, we demonstrate that the mutant can be easily purified in complex with dopamine-bound D2 receptors, and use cryo-electron microscopy to determine the structure, including both domains of Gα, without nucleotide or stabilizing nanobodies. These findings reveal the molecular basis for the first committed step of G protein activation, establish a mechanistic basis for a neurological disorder, provide a simplified strategy to determine receptor-G protein structures, and a method to detect high affinity agonist binding in cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41766.map.gz emd_41766.map.gz | 85.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41766-v30.xml emd-41766-v30.xml emd-41766.xml emd-41766.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41766.png emd_41766.png | 83.6 KB | ||
| Filedesc metadata |  emd-41766.cif.gz emd-41766.cif.gz | 7.3 KB | ||
| Others |  emd_41766_half_map_1.map.gz emd_41766_half_map_1.map.gz emd_41766_half_map_2.map.gz emd_41766_half_map_2.map.gz | 159.7 MB 159.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41766 http://ftp.pdbj.org/pub/emdb/structures/EMD-41766 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41766 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41766 | HTTPS FTP |
-Related structure data
| Related structure data |  8tzqMC  8u02C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41766.map.gz / Format: CCP4 / Size: 172.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41766.map.gz / Format: CCP4 / Size: 172.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.874 Å | ||||||||||||||||||||||||||||||||||||
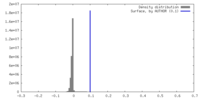
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41766_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
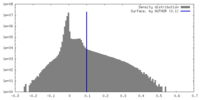
| Density Histograms |
-Half map: #1
| File | emd_41766_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
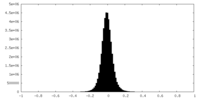
| Density Histograms |
- Sample components
Sample components
-Entire : Human DRD2 in complex with heterotrimeric G protein GoA (K46E), s...
| Entire | Name: Human DRD2 in complex with heterotrimeric G protein GoA (K46E), scFv16, and dopamine |
|---|---|
| Components |
|
-Supramolecule #1: Human DRD2 in complex with heterotrimeric G protein GoA (K46E), s...
| Supramolecule | Name: Human DRD2 in complex with heterotrimeric G protein GoA (K46E), scFv16, and dopamine type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 120 KDa |
-Macromolecule #1: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.668922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAALEVLFQ GPHHHHHHHH |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.418086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: D(2) dopamine receptor
| Macromolecule | Name: D(2) dopamine receptor / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.685355 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDPLNLSWYD DDLERQNWSR PFNGSDGKAD RPHYNYYATL LTLLIAVIVF GNVLVCMAVS REKALQTTTN YLIVSLAVAD LLVATLVMP WVVYLEVVGE WKFSRIHCDI FVTLDVMMCT ASILNLCAIS IDRYTAVAMP MLYNTRYSSK RRVTVMISIV W VLSFTISC ...String: MDPLNLSWYD DDLERQNWSR PFNGSDGKAD RPHYNYYATL LTLLIAVIVF GNVLVCMAVS REKALQTTTN YLIVSLAVAD LLVATLVMP WVVYLEVVGE WKFSRIHCDI FVTLDVMMCT ASILNLCAIS IDRYTAVAMP MLYNTRYSSK RRVTVMISIV W VLSFTISC PLLFGLNNAD QNECIIANPA FVVYSSIVSF YVPFIVTLLV YIKIYIVLRR RRKRVNTKRS SRAFRAHLRA PL KGNCTHP EDMKLCTVIM KSNGSFPVNR RRVEAARRAQ ELEMEMLSST SPPERTRYSP IPPSHHQLTL PDPSHHGLHS TPD SPAKPE KNGHAKDHPK IAKIFEIQTM PNGKTRTSLK TMSRRKLSQQ KEKKATQMLA IVLGVFIICW LPFFITHILN IHCD CNIPP VLYSAFTWLG YVNSAVNPII YTTFNIEFRK AFLKILHC UniProtKB: D(2) dopamine receptor |
-Macromolecule #5: Guanine nucleotide-binding protein G(o) subunit alpha
| Macromolecule | Name: Guanine nucleotide-binding protein G(o) subunit alpha / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.100434 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEER AALERSKAIE KNLKEDGISA AKDVKLLLLG AGESGESTIV KQMKIIHEDG FSGEDVKQYK PVVYSNTIQS LAAIVRAMD TLGIEYGDKE RKADAKMVCD VVSRMEDTEP FSAELLSAMM RLWGDSGIQE CFNRSREYQL NDSAKYYLDS L DRIGAADY ...String: MGCTLSAEER AALERSKAIE KNLKEDGISA AKDVKLLLLG AGESGESTIV KQMKIIHEDG FSGEDVKQYK PVVYSNTIQS LAAIVRAMD TLGIEYGDKE RKADAKMVCD VVSRMEDTEP FSAELLSAMM RLWGDSGIQE CFNRSREYQL NDSAKYYLDS L DRIGAADY QPTEQDILRT RVKTTGIVET HFTFKNLHFR LFDVGGQRSE RKKWIHCFED VTAIIFCVAL SGYDQVLHED ET TNRMHES LMLFDSICNN KFFIDTSIIL FLNKKDLFGE KIKKSPLTIC FPEYTGPNTY EDAAAYIQAQ FESKNRSPNK EIY CHMTCA TDTNNIQVVF DAVTDIIIAN NLRGCGLY UniProtKB: Guanine nucleotide-binding protein G(o) subunit alpha |
-Macromolecule #6: L-DOPAMINE
| Macromolecule | Name: L-DOPAMINE / type: ligand / ID: 6 / Number of copies: 1 / Formula: LDP |
|---|---|
| Molecular weight | Theoretical: 153.178 Da |
| Chemical component information |  ChemComp-LDP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)