-Search query
-Search result
Showing 1 - 50 of 52 items for (author: white & jm)

EMDB-51930: 
BAM-hinge (LVPR)
Method: single particle / : Machin JM, Ranson NA

EMDB-51931: 
BAM-hinge (GSGS)
Method: single particle / : Machin JM, Ranson NA

EMDB-51933: 
BAM-hinge (LVPR) suppressor (T434A)
Method: single particle / : Machin JM, Ranson NA

EMDB-43250: 
SARS-CoV-2 spike omicron (BA.1) ectodomain trimer in complex with SC27 Fab, global refinement
Method: single particle / : Byrne PO, McLellan JS

EMDB-43261: 
SARS-CoV-2 spike omicron (BA.1) ectodomain dimer-of-trimers in complex with SC27 Fab, global refinement
Method: single particle / : Byrne PO, McLellan JS

EMDB-43260: 
SARS-CoV-2 spike omicron (BA.1) ectodomain trimer in complex with SC27 Fab, local refinement
Method: single particle / : Byrne PO, McLellan JS

EMDB-43315: 
SARS-CoV-2 spike omicron (BA.1) RBD ectodomain dimer-of-trimers in complex with SC27 Fabs
Method: single particle / : Byrne PO, McLellan JS

EMDB-44635: 
Inactive mu opioid receptor bound to Nb6, naloxone and NAM
Method: single particle / : O'Brien ES, Wang H, Kaavya Krishna K, Zhang C, Kobilka BK

EMDB-40271: 
Mycobacterium phage Adjutor
Method: single particle / : Podgorski JM, White SJ

EMDB-27271: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH

EMDB-27272: 
CryoEM structure of Western equine encephalitis virus VLP
Method: single particle / : Zimmerman MI, Fremont DH

EMDB-40711: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-28761: 
Mycobacterium phage Che8 mutant capsid (gene 110 deletion)
Method: single particle / : Podgorski JM, White SJ

EMDB-28644: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-16268: 
E. coli BAM complex (BamABCDE) bound to darobactin B
Method: single particle / : Horne JE, Fenn KL, Radford SE, Ranson NA

EMDB-16282: 
E. coli BAM complex (BamABCDE) wild-type
Method: single particle / : Machin JM, Radford SE, Ranson NA

EMDB-40077: 
Mycobacterium phage Patience
Method: single particle / : Podgorski JM, White SJ

EMDB-27824: 
Mycobacterium phage Che8
Method: single particle / : Podgorski JM, White SJ

EMDB-27992: 
Gordonia phage Ziko
Method: single particle / : Podgorski JM, White SJ

EMDB-28012: 
Mycobacterium phage Adephagia
Method: single particle / : Podgorski JM, White SJ

EMDB-28015: 
Mycobacterium phage Bobi
Method: single particle / : Podgorski JM, White SJ

EMDB-28016: 
Arthrobacter phage Bridgette
Method: single particle / : Podgorski JM, White SJ

EMDB-28017: 
Mycobacterium phage Cain
Method: single particle / : Podgorski JM, White SJ

EMDB-28018: 
Gordonia phage Cozz
Method: single particle / : Podgorski JM, White SJ

EMDB-28020: 
Mycobacterium phage Ogopogo
Method: single particle / : Podgorski JM, White SJ

EMDB-28021: 
Microbacterium phage Oxtober96
Method: single particle / : Podgorski JM, White SJ

EMDB-28039: 
Mycobacteriophage Muddy capsid
Method: single particle / : Freeman KG, White SJ, Huet A, Conway JF

EMDB-24482: 
Complex III2 from Candida albicans, inhibitor free
Method: single particle / : Di Trani JM, Rubinstein JL

EMDB-24483: 
Complex III2 from Candida albicans, inhibitor free, Rieske head domain in b position
Method: single particle / : Di Trani JM, Rubinstein JL

EMDB-24484: 
Complex III2 from Candida albicans, inhibitor free, Rieske head domain in intermediate position
Method: single particle / : Di Trani JM, Rubinstein JL

EMDB-24485: 
Complex III2 from Candida albicans, inhibitor free, Rieske head domain in c position
Method: single particle / : Di Trani JM, Rubinstein JL

EMDB-24486: 
Complex III2 from Candida albicans, Inz-5 bound
Method: single particle / : Di Trani JM, Rubinstein JL

EMDB-12232: 
Lateral-closed conformation of the lid-locked BAM complex (BamA E435C S665C, BamBDCE) by cryoEM
Method: single particle / : Haysom SF

EMDB-12262: 
Lateral-open conformation of the lid-locked BAM complex (BamA E435C S665C, BamBDCE) by cryoEM
Method: single particle / : Haysom SF

EMDB-12263: 
structure of a POTRA-locked BAM complex in a lateral-open conformation
Method: single particle / : Machin JM, Haysom SF

EMDB-12271: 
Lateral-open conformation of the lid-locked BAM complex (BamA E435C S665C, BamBDCE) bound by a bactericidal Fab fragment
Method: single particle / : Machin JM, Haysom SF

EMDB-12272: 
lateral-open conformation of the wild-type BAM complex (BamABCDE) bound to a bactericidal Fab fragment
Method: single particle / : Iadanza MG

EMDB-22829: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

EMDB-21122: 
Actinobacteriophage Rosebush
Method: single particle / : Podgorski JM, Calabrese J, Alexandrescu L, Jacobs-Sera D, Pope W, Hatfull G, White SJ

EMDB-21123: 
Actinobacteriophage Patience
Method: single particle / : Podgorski JM, Calabrese J, Alexandrescu L, Jacobs-Sera D, Pope W, Hatfull G, White SJ

EMDB-21124: 
Actinobacteriophage Myrna
Method: single particle / : Podgorski JM, Calabrese J, Alexandrescu L, Jacobs-Sera D, Pope W, Hatfull G, White SJ

EMDB-8225: 
Tomographic subvolume average of Ebola (EBOV-Makona) glycoprotein on the surface of virus-like particles
Method: subtomogram averaging / : Tran EEH, Subramaniam S

EMDB-8226: 
Tomographic subvolume average of membrane-bound Ebola (EBOV-Makona) glycoprotein bound to c13C6 antibody
Method: subtomogram averaging / : Tran EEH, Subramaniam S
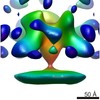
EMDB-8227: 
Tomographic subvolume average of membrane-bound Ebola (EBOV-Makona) glycoprotein bound to c2G4 antibody
Method: subtomogram averaging / : Tran EEH, Subramaniam S

EMDB-8228: 
Tomographic subvolume average of membrane-bound Ebola (EBOV-Makona) glycoproteins bound to the chimerized human monoclonal antibody, c4G7
Method: subtomogram averaging / : Tran EEH, Subramaniam S

EMDB-6003: 
Cryo-electron tomography of full-length glycoprotein from Ebola virus-like particles
Method: subtomogram averaging / : Tran EEH, Simmons JA, Bartesaghi A, Shoemaker CJ, Nelson E, White JM, Subramaniam S

EMDB-6004: 
Cryo-electron tomography of glycoprotein lacking the mucin-like domain from Ebola virus-like particles
Method: subtomogram averaging / : Tran EEH, Simmons JA, Bartesaghi A, Shoemaker CJ, Nelson E, White JM, Subramaniam S
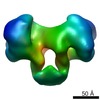
EMDB-1890: 
EcoR124 Type I DNA restriction-modification enzyme complex in closed state with bound 30bp cognate DNA fragment. 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF

EMDB-1891: 
EcoR124 Type I DNA restriction-modification enzyme complex without DNA (open state). Low resolution 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF
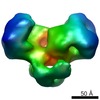
EMDB-1892: 
EcoR124 Type I DNA restriction-modification enzyme complex (in closed state) with bound DNA mimic protein Ocr from phage T7. 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
