+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mycobacteriophage Muddy capsid | |||||||||||||||
 Map data Map data | Sharpened map of Bayesian polished and ewald sphere corrected Muddy_postprocess. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Bacteriophage / Mycobacteriophage / HK97-fold / Capsid / VIRUS | |||||||||||||||
| Function / homology | : / Phage capsid / Phage capsid family / virion component / Capsid protein Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Mycobacterium phage Muddy (virus) Mycobacterium phage Muddy (virus) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||||||||
 Authors Authors | Freeman KG / White SJ / Huet A / Conway JF | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability. Authors: Jennifer M Podgorski / Krista Freeman / Sophia Gosselin / Alexis Huet / James F Conway / Mary Bird / John Grecco / Shreya Patel / Deborah Jacobs-Sera / Graham Hatfull / Johann Peter Gogarten ...Authors: Jennifer M Podgorski / Krista Freeman / Sophia Gosselin / Alexis Huet / James F Conway / Mary Bird / John Grecco / Shreya Patel / Deborah Jacobs-Sera / Graham Hatfull / Johann Peter Gogarten / Janne Ravantti / Simon J White /   Abstract: Many double-stranded DNA viruses, including tailed bacteriophages (phages) and herpesviruses, use the HK97-fold in their major capsid protein to make the capsomers of the icosahedral viral capsid. ...Many double-stranded DNA viruses, including tailed bacteriophages (phages) and herpesviruses, use the HK97-fold in their major capsid protein to make the capsomers of the icosahedral viral capsid. After the genome packaging at near-crystalline densities, the capsid is subjected to a major expansion and stabilization step that allows it to withstand environmental stresses and internal high pressure. Several different mechanisms for stabilizing the capsid have been structurally characterized, but how these mechanisms have evolved is still not understood. Using cryo-EM structure determination of 10 capsids, structural comparisons, phylogenetic analyses, and Alphafold predictions, we have constructed a detailed structural dendrogram describing the evolution of capsid structural stability within the actinobacteriophages. We show that the actinobacteriophage major capsid proteins can be classified into 15 groups based upon their HK97-fold. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28039.map.gz emd_28039.map.gz | 1.8 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28039-v30.xml emd-28039-v30.xml emd-28039.xml emd-28039.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28039_fsc.xml emd_28039_fsc.xml | 28 KB | Display |  FSC data file FSC data file |
| Images |  emd_28039.png emd_28039.png | 322.5 KB | ||
| Masks |  emd_28039_msk_1.map emd_28039_msk_1.map | 1.9 GB |  Mask map Mask map | |
| Filedesc metadata |  emd-28039.cif.gz emd-28039.cif.gz | 6.4 KB | ||
| Others |  emd_28039_additional_1.map.gz emd_28039_additional_1.map.gz emd_28039_additional_2.map.gz emd_28039_additional_2.map.gz emd_28039_half_map_1.map.gz emd_28039_half_map_1.map.gz emd_28039_half_map_2.map.gz emd_28039_half_map_2.map.gz | 1.8 GB 1.5 GB 1.6 GB 1.6 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28039 http://ftp.pdbj.org/pub/emdb/structures/EMD-28039 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28039 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28039 | HTTPS FTP |
-Related structure data
| Related structure data |  8eduMC  8e16C  8eb4C  8ec2C  8ec8C  8eciC  8ecjC  8eckC  8ecnC  8ecoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28039.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28039.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of Bayesian polished and ewald sphere corrected Muddy_postprocess. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.076 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28039_msk_1.map emd_28039_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
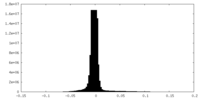
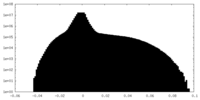
| Density Histograms |
-Additional map: Ewald sphere corrected map of Muddy 3DRefine After Polishing And CTFRefine....
| File | emd_28039_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ewald sphere corrected map of Muddy_3DRefine_After_Polishing_And_CTFRefine. | ||||||||||||
| Projections & Slices |
| ||||||||||||
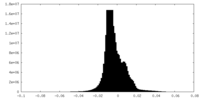
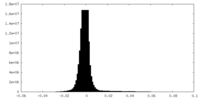
| Density Histograms |
-Additional map: Map before ewald sphere correction but after Bayesian...
| File | emd_28039_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map before ewald sphere correction but after Bayesian polishing and CTF Refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
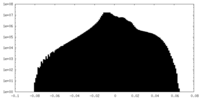
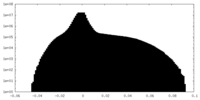
| Density Histograms |
-Half map: Half map of ewald sphere corrected Muddy 3DRefine After Polishing And CTFRefine.mrc....
| File | emd_28039_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of ewald sphere corrected Muddy_3DRefine_After_Polishing_And_CTFRefine.mrc. | ||||||||||||
| Projections & Slices |
| ||||||||||||
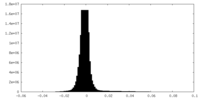
| Density Histograms |
-Half map: Half map of ewald sphere corrected Muddy 3DRefine After Polishing And CTFRefine.mrc....
| File | emd_28039_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of ewald sphere corrected Muddy_3DRefine_After_Polishing_And_CTFRefine.mrc. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mycobacterium phage Muddy
| Entire | Name:  Mycobacterium phage Muddy (virus) Mycobacterium phage Muddy (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Mycobacterium phage Muddy
| Supramolecule | Name: Mycobacterium phage Muddy / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Phage Muddy particles generated by amplification on bacterial host and purification via CsCl gradient. NCBI-ID: 1340829 / Sci species name: Mycobacterium phage Muddy / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Virus shell | Shell ID: 1 / Diameter: 710.0 Å / T number (triangulation number): 7 |
-Macromolecule #1: Capsid
| Macromolecule | Name: Capsid / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium phage Muddy (virus) Mycobacterium phage Muddy (virus) |
| Molecular weight | Theoretical: 34.670102 KDa |
| Sequence | String: AGFANIQGRA DLSDVHLPDQ VIKDVLQTAP EASVLLNRAR KVRMSSKKTK QPVLASLPDA YWVDGDTGLK QTTKNIWSNV FMTAEELAV IVPIPDALIA DSDLPLWDEV KPLLVEAIGK KVDDAGIFGN DKPASWPAAL IPGAIAAGNS VTLGTGDDIG V DVATLGEQ ...String: AGFANIQGRA DLSDVHLPDQ VIKDVLQTAP EASVLLNRAR KVRMSSKKTK QPVLASLPDA YWVDGDTGLK QTTKNIWSNV FMTAEELAV IVPIPDALIA DSDLPLWDEV KPLLVEAIGK KVDDAGIFGN DKPASWPAAL IPGAIAAGNS VTLGTGDDIG V DVATLGEQ LALDGFSING FISRPGLHWS LVGLRNAQGQ PIYTPPLSTG LNGAPPTPAL YGFPLNEVTS GVWDADEAIL LG ADWSKVV IGIRQDITFD LFSEGVISDS DGKVVLNLMQ QDSKALRVVF RVGFQVANPM TRLNPNEATR YPAGVIIPAG GGS GEGEGE SE UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 1027 / Average exposure time: 40.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Amino acid sequence built into the map for a single major capsid protein and refined with Phenix. Model then used for rest of asymmetric unit and refined with Phenix. Final step involved using Isolde. |
|---|---|
| Refinement | Protocol: AB INITIO MODEL |
| Output model |  PDB-8edu: |
 Movie
Movie Controller
Controller















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)