+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21123 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Actinobacteriophage Patience | |||||||||
 Map data Map data | Actinobacteriophage Patience | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Mycobacterium phage Patience (virus) Mycobacterium phage Patience (virus) | |||||||||
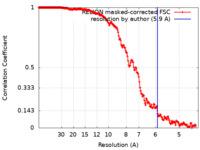
| Method | single particle reconstruction / cryo EM / Resolution: 5.9 Å | |||||||||
 Authors Authors | Podgorski JM / Calabrese J / Alexandrescu L / Jacobs-Sera D / Pope W / Hatfull G / White SJ | |||||||||
 Citation Citation |  Journal: Viruses / Year: 2020 Journal: Viruses / Year: 2020Title: Structures of Three Actinobacteriophage Capsids: Roles of Symmetry and Accessory Proteins. Authors: Jennifer Podgorski / Joshua Calabrese / Lauren Alexandrescu / Deborah Jacobs-Sera / Welkin Pope / Graham Hatfull / Simon White /  Abstract: Here, we describe the structure of three actinobacteriophage capsids that infect . The capsid structures were resolved to approximately six angstroms, which allowed confirmation that each ...Here, we describe the structure of three actinobacteriophage capsids that infect . The capsid structures were resolved to approximately six angstroms, which allowed confirmation that each bacteriophage uses the HK97-fold to form their capsid. One bacteriophage, Rosebush, may have a novel variation of the HK97-fold. Four novel accessory proteins that form the capsid head along with the major capsid protein were identified. Two of the accessory proteins were minor capsid proteins and showed some homology, based on bioinformatic analysis, to the TW1 bacteriophage. The remaining two accessory proteins are decoration proteins that are located on the outside of the capsid and do not resemble any previously described bacteriophage decoration protein. SDS-PAGE and mass spectrometry was used to identify the accessory proteins and bioinformatic analysis of the accessory proteins suggest they are used in many actinobacteriophage capsids. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21123.map.gz emd_21123.map.gz | 220.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21123-v30.xml emd-21123-v30.xml emd-21123.xml emd-21123.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21123_fsc.xml emd_21123_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_21123.png emd_21123.png | 92 KB | ||
| Masks |  emd_21123_msk_1.map emd_21123_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Others |  emd_21123_additional.map.gz emd_21123_additional.map.gz emd_21123_additional_1.map.gz emd_21123_additional_1.map.gz emd_21123_half_map_1.map.gz emd_21123_half_map_1.map.gz emd_21123_half_map_2.map.gz emd_21123_half_map_2.map.gz | 190.5 MB 190.5 MB 94.4 MB 94.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21123 http://ftp.pdbj.org/pub/emdb/structures/EMD-21123 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21123 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21123 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21123.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21123.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Actinobacteriophage Patience | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
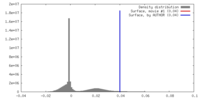
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21123_msk_1.map emd_21123_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
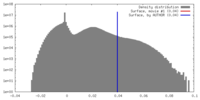
| Density Histograms |
-Additional map: Unpolished map
| File | emd_21123_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unpolished map | ||||||||||||
| Projections & Slices |
| ||||||||||||
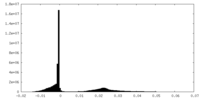
| Density Histograms |
-Additional map: Unpolished map
| File | emd_21123_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unpolished map | ||||||||||||
| Projections & Slices |
| ||||||||||||
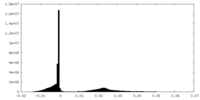
| Density Histograms |
-Half map: #2
| File | emd_21123_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_21123_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mycobacterium phage Patience
| Entire | Name:  Mycobacterium phage Patience (virus) Mycobacterium phage Patience (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Mycobacterium phage Patience
| Supramolecule | Name: Mycobacterium phage Patience / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Grown in host Mycobacterium smegmatis and cesium chloride-purified NCBI-ID: 1074308 / Sci species name: Mycobacterium phage Patience / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Virus shell | Shell ID: 1 / T number (triangulation number): 7 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Support film - Material: CARBON / Support film - topology: HOLEY / Details: unspecified | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | This sample was multiplexed with two other bacteriophages. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 996 / Average exposure time: 1.6 sec. / Average electron dose: 17.6 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)