-Search query
-Search result
Showing 1 - 50 of 66 items for (author: tobiasson & v)

EMDB-17045: 
CryoEM structure of human rho1 GABAA receptor in complex with GABA
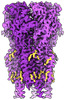
EMDB-17106: 
CryoEM structure of human rho1 GABAA receptor apo state

EMDB-17107: 
CryoEM structure of human rho1 GABAA receptor in complex with inhibitor TPMPA
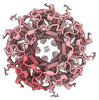
EMDB-17108: 
CryoEM structure of human rho1 GABAA receptor in complex with pore blocker picrotoxin
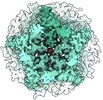
EMDB-17110: 
CryoEM structure of human rho1 GABAA receptor in complex with GABA and picrotoxin

PDB-8op9: 
CryoEM structure of human rho1 GABAA receptor in complex with GABA

PDB-8oq6: 
CryoEM structure of human rho1 GABAA receptor apo state

PDB-8oq7: 
CryoEM structure of human rho1 GABAA receptor in complex with inhibitor TPMPA

PDB-8oq8: 
CryoEM structure of human rho1 GABAA receptor in complex with pore blocker picrotoxin

PDB-8oqa: 
CryoEM structure of human rho1 GABAA receptor in complex with GABA and picrotoxin

EMDB-16184: 
Cryo-EM structure of the I-II-III2-IV2 respiratory supercomplex from Tetrahymena thermophila

EMDB-15577: 
Structure of the mitochondrial ribosome from Polytomella magna with tRNAs bound to the A and P sites

PDB-8apo: 
Structure of the mitochondrial ribosome from Polytomella magna with tRNAs bound to the A and P sites

EMDB-15900: 
Subtomogram average of the respiratory supercomplex from Tetrahymena thermophila mitochondria

EMDB-15865: 
Cryo-EM structure of NADH:ubiquinone oxidoreductase (complex-I) from respiratory supercomplex of Tetrahymena thermophila

EMDB-15866: 
Cryo-EM structure of succinate dehydrogenase complex (complex-II) in respiratory supercomplex of Tetrahymena thermophila

EMDB-15867: 
Cryo-EM structure of cytochrome c oxidase dimer (complex IV) from respiratory supercomplex of Tetrahymena thermophila

EMDB-15868: 
Cryo-EM structure of cytochrome bc1 complex (complex-III) from respiratory supercomplex of Tetrahymena thermophila

PDB-8b6f: 
Cryo-EM structure of NADH:ubiquinone oxidoreductase (complex-I) from respiratory supercomplex of Tetrahymena thermophila

PDB-8b6g: 
Cryo-EM structure of succinate dehydrogenase complex (complex-II) in respiratory supercomplex of Tetrahymena thermophila

PDB-8b6h: 
Cryo-EM structure of cytochrome c oxidase dimer (complex IV) from respiratory supercomplex of Tetrahymena thermophila

PDB-8b6j: 
Cryo-EM structure of cytochrome bc1 complex (complex-III) from respiratory supercomplex of Tetrahymena thermophila

EMDB-14867: 
Dimeric PSI of Chlamydomonas reinhardtii at 2.74 A resolution (symmetry expanded)

EMDB-14870: 
Monomeric PSI of Chlamydomonas reinhardtii at 2.31 A resolution

EMDB-14871: 
Dimeric PSI of Chlamydomonas reinhardtii at 2.97 A resolution

EMDB-14872: 
Plastocyanin bound to PSI of Chlamydomonas reinhardtii

PDB-7zq9: 
Dimeric PSI of Chlamydomonas reinhardtii at 2.74 A resolution (symmetry expanded)

PDB-7zqc: 
Monomeric PSI of Chlamydomonas reinhardtii at 2.31 A resolution

PDB-7zqd: 
Dimeric PSI of Chlamydomonas reinhardtii at 2.97 A resolution

PDB-7zqe: 
Plastocyanin bound to PSI of Chlamydomonas reinhardtii

EMDB-15100: 
Structure of the mitochondrial ribosome from Polytomella magna

EMDB-15576: 
Structure of the mitochondrial ribosome from Polytomella magna with tRNA bound to the P site

PDB-8a22: 
Structure of the mitochondrial ribosome from Polytomella magna

PDB-8apn: 
Structure of the mitochondrial ribosome from Polytomella magna with tRNA bound to the P site

EMDB-11845: 
Trypanosoma brucei mitochondrial ribosome large subunit assembly intermediate

PDB-7aoi: 
Trypanosoma brucei mitochondrial ribosome large subunit assembly intermediate

EMDB-10857: 
Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - Fo-wing region

EMDB-10858: 
Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - central stalk/cring

EMDB-10859: 
Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - Fo-subcomplex

EMDB-10860: 
Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1Fo dimer

EMDB-10861: 
Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1Fo tetramer

EMDB-10862: 
Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1/peripheral stalk

PDB-6ynv: 
Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - Fo-wing region

PDB-6ynw: 
Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - central stalk/cring

PDB-6ynx: 
Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - Fo-subcomplex

PDB-6yny: 
Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1Fo composite dimer model

PDB-6ynz: 
Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1Fo composite tetramer model

PDB-6yo0: 
Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1/peripheral stalk

EMDB-11033: 
The mitochondrial ribosome from Tetrahymena thermophila, large subunit mask

EMDB-11034: 
The mitochondrial ribosome from Tetrahymena thermophila, small subunit mask
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
