[English] 日本語
 Yorodumi
Yorodumi- EMDB-15865: Cryo-EM structure of NADH:ubiquinone oxidoreductase (complex-I) f... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of NADH:ubiquinone oxidoreductase (complex-I) from respiratory supercomplex of Tetrahymena thermophila | |||||||||||||||
 Map data Map data | Sharpened mask refined map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Ciliate / mitochondrial / complex-I / supercomplex / ELECTRON TRANSPORT | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationlipid-A-disaccharide synthase / lipid-A-disaccharide synthase activity / NADH:ubiquinone reductase (H+-translocating) / membrane protein complex / P450-containing electron transport chain / NADH dehydrogenase complex / oxidoreductase activity, acting on NAD(P)H / lipid A biosynthetic process / ligase activity / acyl binding ...lipid-A-disaccharide synthase / lipid-A-disaccharide synthase activity / NADH:ubiquinone reductase (H+-translocating) / membrane protein complex / P450-containing electron transport chain / NADH dehydrogenase complex / oxidoreductase activity, acting on NAD(P)H / lipid A biosynthetic process / ligase activity / acyl binding / electron transport coupled proton transport / NADH:ubiquinone reductase (H+-translocating) / acyl carrier activity / NADH dehydrogenase activity / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / respiratory chain complex I / NADH dehydrogenase (ubiquinone) activity / catalytic complex / quinone binding / ATP synthesis coupled electron transport / protein folding chaperone / aerobic respiration / respiratory electron transport chain / electron transport chain / phospholipid binding / 2 iron, 2 sulfur cluster binding / mitochondrial membrane / NAD binding / unfolded protein binding / FMN binding / protein-folding chaperone binding / 4 iron, 4 sulfur cluster binding / response to oxidative stress / electron transfer activity / oxidoreductase activity / mitochondrial inner membrane / ribosome / protein-containing complex binding / mitochondrion / zinc ion binding / metal ion binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |   Tetrahymena thermophila SB210 (eukaryote) Tetrahymena thermophila SB210 (eukaryote) | |||||||||||||||
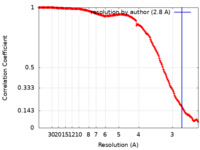
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||||||||
 Authors Authors | Muhleip A / Kock Flygaard R / Amunts A | |||||||||||||||
| Funding support |  Sweden, European Union, 4 items Sweden, European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structural basis of mitochondrial membrane bending by the I-II-III-IV supercomplex. Authors: Alexander Mühleip / Rasmus Kock Flygaard / Rozbeh Baradaran / Outi Haapanen / Thomas Gruhl / Victor Tobiasson / Amandine Maréchal / Vivek Sharma / Alexey Amunts /      Abstract: Mitochondrial energy conversion requires an intricate architecture of the inner mitochondrial membrane. Here we show that a supercomplex containing all four respiratory chain components contributes ...Mitochondrial energy conversion requires an intricate architecture of the inner mitochondrial membrane. Here we show that a supercomplex containing all four respiratory chain components contributes to membrane curvature induction in ciliates. We report cryo-electron microscopy and cryo-tomography structures of the supercomplex that comprises 150 different proteins and 311 bound lipids, forming a stable 5.8-MDa assembly. Owing to subunit acquisition and extension, complex I associates with a complex IV dimer, generating a wedge-shaped gap that serves as a binding site for complex II. Together with a tilted complex III dimer association, it results in a curved membrane region. Using molecular dynamics simulations, we demonstrate that the divergent supercomplex actively contributes to the membrane curvature induction and tubulation of cristae. Our findings highlight how the evolution of protein subunits of respiratory complexes has led to the I-II-III-IV supercomplex that contributes to the shaping of the bioenergetic membrane, thereby enabling its functional specialization. #1:  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: Structural basis of mitochondrial membrane bending by I-II-III2-IV2 supercomplex Authors: Muhleip A / Flygaard RK / Haapanen O / Baradaran R / Gruhl T / Tobiasson V / Marechal A / Sharma V / Amunts A | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15865.map.gz emd_15865.map.gz | 398.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15865-v30.xml emd-15865-v30.xml emd-15865.xml emd-15865.xml | 97.2 KB 97.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15865_fsc.xml emd_15865_fsc.xml | 16.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_15865.png emd_15865.png | 117.7 KB | ||
| Masks |  emd_15865_msk_1.map emd_15865_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15865.cif.gz emd-15865.cif.gz | 19.2 KB | ||
| Others |  emd_15865_additional_1.map.gz emd_15865_additional_1.map.gz emd_15865_half_map_1.map.gz emd_15865_half_map_1.map.gz emd_15865_half_map_2.map.gz emd_15865_half_map_2.map.gz | 213.6 MB 391 MB 391 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15865 http://ftp.pdbj.org/pub/emdb/structures/EMD-15865 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15865 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15865 | HTTPS FTP |
-Related structure data
| Related structure data |  8b6fMC  8b6gC  8b6hC  8b6jC  8bqsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15865.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15865.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened mask refined map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.25 Å | ||||||||||||||||||||||||||||||||||||
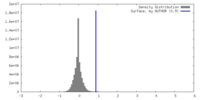
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15865_msk_1.map emd_15865_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
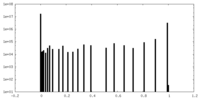
| Projections & Slices |
| ||||||||||||
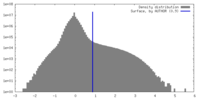
| Density Histograms |
-Additional map: Unsharpened mask refined map
| File | emd_15865_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened mask refined map | ||||||||||||
| Projections & Slices |
| ||||||||||||
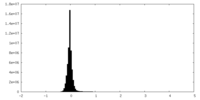
| Density Histograms |
-Half map: Half-map A
| File | emd_15865_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map B
| File | emd_15865_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : NADH:ubiquinone oxidoreductase complex (complex-I) from respirato...
+Supramolecule #1: NADH:ubiquinone oxidoreductase complex (complex-I) from respirato...
+Macromolecule #1: Lipid-A-disaccharide synthase
+Macromolecule #2: NAD-dependent epimerase/dehydratase family protein
+Macromolecule #3: DnaJ domain protein
+Macromolecule #4: Acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II
+Macromolecule #5: RNase III domain-containing protein
+Macromolecule #6: 37S ribosomal protein S25, mitochondrial
+Macromolecule #7: Transmembrane protein, putative
+Macromolecule #8: CX9C domain-containing protein
+Macromolecule #9: NDUTT15
+Macromolecule #10: Transmembrane protein, putative
+Macromolecule #11: NADH dehydrogenase subunit 5
+Macromolecule #12: NADH-ubiquinone oxidoreductase 75 kDa subunit
+Macromolecule #13: NADH-ubiquinone oxidoreductase chain 4
+Macromolecule #14: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial
+Macromolecule #15: NADH dehydrogenase subunit 7
+Macromolecule #16: Ymf65
+Macromolecule #17: Transcription factor apfi protein, putative
+Macromolecule #18: NADH-ubiquinone oxidoreductase chain 1
+Macromolecule #19: NADH-ubiquinone oxidoreductase 24 kDa subunit
+Macromolecule #20: Ymf62
+Macromolecule #21: Gamma-carbonic anhydrase
+Macromolecule #22: NADH-ubiquinone oxidoreductase 1, chain, putative
+Macromolecule #23: Gamma-carbonic anhydrase
+Macromolecule #24: ETC complex I subunit motif protein
+Macromolecule #25: NADH dehydrogenase subunit 9
+Macromolecule #26: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12
+Macromolecule #27: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mi...
+Macromolecule #28: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial
+Macromolecule #29: NADH dehydrogenase, putative
+Macromolecule #30: NADH dehydrogenase subunit 10
+Macromolecule #31: NADH-ubiquinone oxidoreductase complex I, 21 kDa subunit
+Macromolecule #32: Acyl carrier protein
+Macromolecule #33: Acyl carrier protein
+Macromolecule #34: NADH-ubiquinone oxidoreductase chain 3
+Macromolecule #35: Ymf58
+Macromolecule #36: Ribosomal protein L51/S25/CI-B8 domain protein
+Macromolecule #37: Transmembrane protein, putative
+Macromolecule #38: ATP synthase subunit e, mitochondrial
+Macromolecule #39: GRAM domain protein
+Macromolecule #40: Transmembrane protein, putative
+Macromolecule #41: Transmembrane protein, putative
+Macromolecule #42: Transmembrane protein, putative
+Macromolecule #43: ND1b
+Macromolecule #44: Transmembrane protein
+Macromolecule #45: Transmembrane protein, putative
+Macromolecule #46: NDUB8
+Macromolecule #47: Transmembrane protein, putative
+Macromolecule #48: NDUPH2
+Macromolecule #49: NDUB10
+Macromolecule #50: NDUA13
+Macromolecule #51: NADH dehydrogenase subunit 2
+Macromolecule #52: 2 iron, 2 sulfur cluster-binding protein
+Macromolecule #53: Thioredoxin
+Macromolecule #54: COX assembly mitochondrial protein
+Macromolecule #55: Transmembrane protein, putative
+Macromolecule #56: Transmembrane protein, putative
+Macromolecule #57: PH domain-containing protein
+Macromolecule #58: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8
+Macromolecule #59: NDUB6
+Macromolecule #60: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4
+Macromolecule #61: Zinc-finger protein
+Macromolecule #62: NDUB4
+Macromolecule #63: NDUB15
+Macromolecule #64: NDUTT16
+Macromolecule #65: NDUTT17
+Macromolecule #66: CHCH domain-containing protein
+Macromolecule #67: Transmembrane protein, putative
+Macromolecule #68: Ymf57
+Macromolecule #69: Complex I-MNLL
+Macromolecule #70: CARDIOLIPIN
+Macromolecule #71: URIDINE-5'-DIPHOSPHATE
+Macromolecule #72: MAGNESIUM ION
+Macromolecule #73: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+Macromolecule #74: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
+Macromolecule #75: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+Macromolecule #76: 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE
+Macromolecule #77: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #78: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #79: IRON/SULFUR CLUSTER
+Macromolecule #80: FLAVIN MONONUCLEOTIDE
+Macromolecule #81: S-[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-b...
+Macromolecule #82: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 25.66 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)