[English] 日本語
 Yorodumi
Yorodumi- EMDB-10860: Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP sy... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10860 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1Fo dimer | ||||||||||||
 Map data Map data | Local-resolution filtered full map of T. thermophila ATP synthase | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | mitochondria / ATP synthase / F1Fo dimer / IF1 dimer / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology information: / sulfide oxidation, using sulfide:quinone oxidoreductase / sulfide:quinone oxidoreductase activity / carboxylic ester hydrolase activity / proton transmembrane transporter activity / proton motive force-driven ATP synthesis / proton-transporting two-sector ATPase complex, proton-transporting domain / proton motive force-driven mitochondrial ATP synthesis / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex ...: / sulfide oxidation, using sulfide:quinone oxidoreductase / sulfide:quinone oxidoreductase activity / carboxylic ester hydrolase activity / proton transmembrane transporter activity / proton motive force-driven ATP synthesis / proton-transporting two-sector ATPase complex, proton-transporting domain / proton motive force-driven mitochondrial ATP synthesis / H+-transporting two-sector ATPase / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / FAD binding / ADP binding / mitochondrial inner membrane / hydrolase activity / lipid binding / mitochondrion / ATP binding / membrane Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
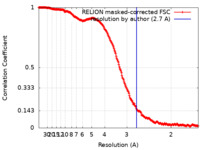
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | ||||||||||||
 Authors Authors | Kock Flygaard R / Muhleip A | ||||||||||||
| Funding support |  Sweden, 3 items Sweden, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization. Authors: Rasmus Kock Flygaard / Alexander Mühleip / Victor Tobiasson / Alexey Amunts /  Abstract: Mitochondrial ATP synthases form functional homodimers to induce cristae curvature that is a universal property of mitochondria. To expand on the understanding of this fundamental phenomenon, we ...Mitochondrial ATP synthases form functional homodimers to induce cristae curvature that is a universal property of mitochondria. To expand on the understanding of this fundamental phenomenon, we characterized the unique type III mitochondrial ATP synthase in its dimeric and tetrameric form. The cryo-EM structure of a ciliate ATP synthase dimer reveals an unusual U-shaped assembly of 81 proteins, including a substoichiometrically bound ATPTT2, 40 lipids, and co-factors NAD and CoQ. A single copy of subunit ATPTT2 functions as a membrane anchor for the dimeric inhibitor IF. Type III specific linker proteins stably tie the ATP synthase monomers in parallel to each other. The intricate dimer architecture is scaffolded by an extended subunit-a that provides a template for both intra- and inter-dimer interactions. The latter results in the formation of tetramer assemblies, the membrane part of which we determined to 3.1 Å resolution. The structure of the type III ATP synthase tetramer and its associated lipids suggests that it is the intact unit propagating the membrane curvature. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10860.map.gz emd_10860.map.gz | 473.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10860-v30.xml emd-10860-v30.xml emd-10860.xml emd-10860.xml | 56.4 KB 56.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10860_fsc.xml emd_10860_fsc.xml | 21.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_10860.png emd_10860.png | 81.6 KB | ||
| Masks |  emd_10860_msk_1.map emd_10860_msk_1.map | 824 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10860.cif.gz emd-10860.cif.gz | 13.2 KB | ||
| Others |  emd_10860_half_map_1.map.gz emd_10860_half_map_1.map.gz emd_10860_half_map_2.map.gz emd_10860_half_map_2.map.gz | 673.3 MB 670.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10860 http://ftp.pdbj.org/pub/emdb/structures/EMD-10860 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10860 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10860 | HTTPS FTP |
-Related structure data
| Related structure data |  6ynyMC  6ynvC  6ynwC  6ynxC  6ynzC  6yo0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10860.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10860.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local-resolution filtered full map of T. thermophila ATP synthase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10860_msk_1.map emd_10860_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1
| File | emd_10860_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2
| File | emd_10860_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Mitochondrial ATP synthase, F1Fo dimer
+Supramolecule #1: Mitochondrial ATP synthase, F1Fo dimer
+Macromolecule #1: subunit a
+Macromolecule #2: subunit b
+Macromolecule #3: subunit d
+Macromolecule #4: subunit f
+Macromolecule #5: subunit i/j
+Macromolecule #6: subunit k
+Macromolecule #7: subunit 8
+Macromolecule #8: ATPTT3
+Macromolecule #9: ATPTT4
+Macromolecule #10: ATPTT5
+Macromolecule #11: ATPTT6
+Macromolecule #12: ATPTT7
+Macromolecule #13: ATPTT8
+Macromolecule #14: ATPTT9
+Macromolecule #15: ATPTT10
+Macromolecule #16: ATPTT11
+Macromolecule #17: ATPTT12
+Macromolecule #18: ATPTT13
+Macromolecule #19: ATPTT1
+Macromolecule #20: Inhibitor of F1 (IF1)
+Macromolecule #21: ATPTT2
+Macromolecule #22: Oligomycin sensitivity-conferring protein (OSCP)
+Macromolecule #23: subunit gamma
+Macromolecule #24: subunit alpha
+Macromolecule #25: subunit beta
+Macromolecule #26: subunit c
+Macromolecule #27: subunit delta
+Macromolecule #28: subunit epsilon
+Macromolecule #29: CARDIOLIPIN
+Macromolecule #30: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+Macromolecule #31: PHOSPHATE ION
+Macromolecule #32: Ubiquinone-8
+Macromolecule #33: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #34: MAGNESIUM ION
+Macromolecule #35: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
+Macromolecule #36: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
+Macromolecule #37: ADENOSINE-5'-DIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.75 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 30.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 165000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)