-Search query
-Search result
Showing all 38 items for (author: thomas & gj)

EMDB-41374: 
Antibody N3-1 bound to RBDs in the up and down conformations
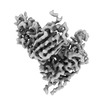
EMDB-41382: 
Antibody N3-1 bound to RBD in the up conformation
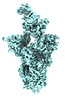
EMDB-41399: 
Antibody N3-1 bound to SARS-CoV-2 spike

PDB-8tm1: 
Antibody N3-1 bound to RBDs in the up and down conformations

PDB-8tma: 
Antibody N3-1 bound to RBD in the up conformation

EMDB-28178: 
Structure of lineage IV Lassa virus glycoprotein complex (strain Josiah)

EMDB-28179: 
Structure of lineage II Lassa virus glycoprotein complex (strain NIG08-A41)

EMDB-28180: 
Structure of lineage V Lassa virus glycoprotein complex (strain Soromba-R)

EMDB-28181: 
Structure of lineage VII Lassa virus glycoprotein complex (strain Togo/2016/7082)

EMDB-28182: 
Lassa virus glycoprotein complex (Josiah) bound to 12.1F Fab

EMDB-28183: 
Lassa virus glycoprotein complex (Josiah) bound to 19.7E Fab

EMDB-28184: 
Lassa virus glycoprotein complex (Josiah) bound to S370.7 Fab
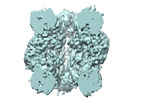
EMDB-27654: 
A subtomogram average of H. neapolitanus Rubisco within alpha-carboxysomes

EMDB-25824: 
aRML prion fibril

EMDB-11928: 
SctV (SsaV) cytoplasmic domain

EMDB-22975: 
Lethocerus Myosin II complete coiled-coil domain resolved in its native environment

PDB-7kog: 
Lethocerus Myosin II complete coiled-coil domain resolved in its native environment

PDB-7beq: 
MicroED structure of the MyD88 TIR domain higher-order assembly

EMDB-20926: 
Clostridium difficile binary toxin translocase CDTb in asymmetric tetradecamer conformation

EMDB-20927: 
Clostridium difficile binary toxin translocase CDTb tetradecamer in symmetric conformation

PDB-6uwr: 
Clostridium difficile binary toxin translocase CDTb in asymmetric tetradecamer conformation

PDB-6uwt: 
Clostridium difficile binary toxin translocase CDTb tetradecamer in symmetric conformation

EMDB-0436: 
Near-atomic structure of icosahedrally averaged PBCV-1 capsid

PDB-6ncl: 
Near-atomic structure of icosahedrally averaged PBCV-1 capsid

EMDB-9004: 
Cryo-EM structure of C. elegans GDP-microtubule

EMDB-7459: 
Cryo-EM structure at 3.8 A resolution of vaccine-elicited antibody vFP20.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122

EMDB-7460: 
Cryo-EM structure at 3.6 A resolution of vaccine-elicited antibody vFP16.02 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122

PDB-6cde: 
Cryo-EM structure at 3.8 A resolution of vaccine-elicited antibody vFP20.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122

PDB-6cdi: 
Cryo-EM structure at 3.6 A resolution of vaccine-elicited antibody vFP16.02 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122

EMDB-8420: 
Cryo-EM structure of BG505 DS-SOSIP HIV-1 Env trimer in complex with vaccine elicited, fusion peptide-directed antibody vFP1.01

EMDB-8421: 
Cryo-EM structure of an asymmetric complex of BG505 DS-SOSIP HIV-1 Env trimer with vaccine elicited, fusion peptide-directed antibody vFP5.01

EMDB-8422: 
Cryo-EM structure of an asymmetric complex of BG505 DS-SOSIP HIV-1 Env trimer with vaccine elicited, fusion peptide-directed antibody vFP5.01

EMDB-2170: 
Thermococcus kodakaraensis 70S ribosome

EMDB-2171: 
Staphylothermus marinus 50S ribosomal subunit

EMDB-2172: 
Methanococcus igneus 70S ribosome

EMDB-1796: 
Proteomic characterization of archaeal ribosomes reveals the presence of novel archaeal-specific ribosomal proteins

EMDB-1797: 
Proteomic characterization of archaeal ribosomes reveals the presence of novel archaeal-specific ribosomal proteins. 50S ribosomal subunit of Sulfolobus acidocaldarius

EMDB-1240: 
The structure of a filamentous bacteriophage.
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
