+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Lassa virus glycoprotein complex (Josiah) bound to S370.7 Fab | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glycoprotein complex / Lassa mammarenavirus / LASV / GPC / immune system / viral fusion protein / Lassa virus / lineage IV / Josiah / S370.7 / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell Golgi membrane / receptor-mediated endocytosis of virus by host cell / host cell endoplasmic reticulum membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Lassa mammarenavirus / Lassa mammarenavirus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
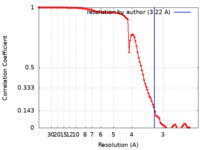
| Method | single particle reconstruction / cryo EM / Resolution: 3.22 Å | |||||||||
 Authors Authors | Perrett HR / Ward AB | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2023 Journal: Cell Rep / Year: 2023Title: Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies. Authors: Hailee R Perrett / Philip J M Brouwer / Jonathan Hurtado / Maddy L Newby / Lin Liu / Helena Müller-Kräuter / Sarah Müller Aguirre / Judith A Burger / Joey H Bouhuijs / Grace Gibson / ...Authors: Hailee R Perrett / Philip J M Brouwer / Jonathan Hurtado / Maddy L Newby / Lin Liu / Helena Müller-Kräuter / Sarah Müller Aguirre / Judith A Burger / Joey H Bouhuijs / Grace Gibson / Terrence Messmer / John S Schieffelin / Aleksandar Antanasijevic / Geert-Jan Boons / Thomas Strecker / Max Crispin / Rogier W Sanders / Bryan Briney / Andrew B Ward /     Abstract: Lassa fever is an acute hemorrhagic fever caused by the zoonotic Lassa virus (LASV). The LASV glycoprotein complex (GPC) mediates viral entry and is the sole target for neutralizing antibodies. ...Lassa fever is an acute hemorrhagic fever caused by the zoonotic Lassa virus (LASV). The LASV glycoprotein complex (GPC) mediates viral entry and is the sole target for neutralizing antibodies. Immunogen design is complicated by the metastable nature of recombinant GPCs and the antigenic differences among phylogenetically distinct LASV lineages. Despite the sequence diversity of the GPC, structures of most lineages are lacking. We present the development and characterization of prefusion-stabilized, trimeric GPCs of LASV lineages II, V, and VII, revealing structural conservation despite sequence diversity. High-resolution structures and biophysical characterization of the GPC in complex with GP1-A-specific antibodies suggest their neutralization mechanisms. Finally, we present the isolation and characterization of a trimer-preferring neutralizing antibody belonging to the GPC-B competition group with an epitope that spans adjacent protomers and includes the fusion peptide. Our work provides molecular detail information on LASV antigenic diversity and will guide efforts to design pan-LASV vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28184.map.gz emd_28184.map.gz | 45.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28184-v30.xml emd-28184-v30.xml emd-28184.xml emd-28184.xml | 27.2 KB 27.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28184_fsc.xml emd_28184_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_28184.png emd_28184.png | 131.7 KB | ||
| Masks |  emd_28184_msk_1.map emd_28184_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-28184.cif.gz emd-28184.cif.gz | 7.7 KB | ||
| Others |  emd_28184_additional_1.map.gz emd_28184_additional_1.map.gz emd_28184_half_map_1.map.gz emd_28184_half_map_1.map.gz emd_28184_half_map_2.map.gz emd_28184_half_map_2.map.gz | 44.5 MB 65.7 MB 65.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28184 http://ftp.pdbj.org/pub/emdb/structures/EMD-28184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28184 | HTTPS FTP |
-Related structure data
| Related structure data |  8ejjMC  8ejdC  8ejeC  8ejfC  8ejgC  8ejhC  8ejiC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28184.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28184.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||
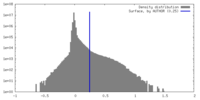
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28184_msk_1.map emd_28184_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
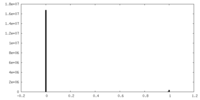
| Density Histograms |
-Additional map: #1
| File | emd_28184_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
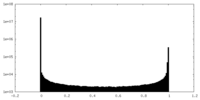
| Density Histograms |
-Half map: #2
| File | emd_28184_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
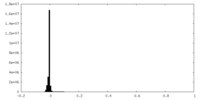
| Density Histograms |
-Half map: #1
| File | emd_28184_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Lassa mammarenavirus GPC
| Entire | Name: Lassa mammarenavirus GPC |
|---|---|
| Components |
|
-Supramolecule #1: Lassa mammarenavirus GPC
| Supramolecule | Name: Lassa mammarenavirus GPC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: GPC protein codon-optimized and expressed in HEK 293F cells using covalently-linked I53-50A trimerization scaffold bound to S370.7 Fab |
|---|---|
| Source (natural) | Organism:  Lassa mammarenavirus / Strain: Josiah Lassa mammarenavirus / Strain: Josiah |
| Molecular weight | Theoretical: 425 KDa |
-Macromolecule #1: Glycoprotein GP1
| Macromolecule | Name: Glycoprotein GP1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lassa mammarenavirus Lassa mammarenavirus |
| Molecular weight | Theoretical: 22.199053 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: TSLYKGVYEL QTLELNMETL NMTMPLSCTK NNSHHYIMVG NETGLELTLT NTSIINHKFC NLSDAHKKNL YDHALMSIIS TFHLSIPNF NQYEAMSCDF NGGKISVQYN LSHSYAGDAA NHCGTVANGV LQTFMRMAWG GSYIALDSGC GNWDCIMTSY Q YLIIQNTT ...String: TSLYKGVYEL QTLELNMETL NMTMPLSCTK NNSHHYIMVG NETGLELTLT NTSIINHKFC NLSDAHKKNL YDHALMSIIS TFHLSIPNF NQYEAMSCDF NGGKISVQYN LSHSYAGDAA NHCGTVANGV LQTFMRMAWG GSYIALDSGC GNWDCIMTSY Q YLIIQNTT WEDHCQFSRP SPIGYLGLLS QRTRDIYIS UniProtKB: Pre-glycoprotein polyprotein GP complex |
-Macromolecule #2: S370.7 heavy chain Fab
| Macromolecule | Name: S370.7 heavy chain Fab / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.109716 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVHLQQWGTG LLKPSETLSL TCAVYGGSIS SYYWSWIRQP PGKGLEWIGE ISHSGSTNYN PSLKSRVTIS VDTSKNQLSL KLRSVTAAD TAVYYCARGR DCRSVSCYVS SPENWFDPWG QGTPITVSS |
-Macromolecule #3: S370.7 light chain Fab
| Macromolecule | Name: S370.7 light chain Fab / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.517611 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: YELTQPPSVS VSPGQTARIT CSGDALPKQY AYWYQQKPGQ APVMVIYKDS ERPSGIPERF SGSNSGTTVT LTISGVQAED EADYYCQSA DSDGTYRVFG GGTKVTVL |
-Macromolecule #4: Glycoprotein GP2
| Macromolecule | Name: Glycoprotein GP2 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lassa mammarenavirus Lassa mammarenavirus |
| Molecular weight | Theoretical: 19.146824 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GTFTWTLSDS EGKDTPGGYC LTRWMLIEAE LKCFGNTAVA KCNEKHDEEF CDMLRLFDFN KQAIQRLKAP AQMSIQLINK AVNALINDQ LIMKNHLRDI MCIPYCNYSK YWYLNHTTTG RTSLPKCWLV SNGSYLNETH FSDDIEQQAD NMITEMLQKE Y MERQ UniProtKB: Pre-glycoprotein polyprotein GP complex |
-Macromolecule #11: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 11 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #12: alpha-D-mannopyranose
| Macromolecule | Name: alpha-D-mannopyranose / type: ligand / ID: 12 / Number of copies: 1 / Formula: MAN |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-MAN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: TBS |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Wait time 10 s; blotting time varied between 3-7 s; blotting force of 0. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.11 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)