-Search query
-Search result
Showing 1 - 50 of 218 items for (author: teo & ew)

EMDB-42118: 
Toxoplasma gondii apical end with ICMAP1 conditionally knocked down

EMDB-42119: 
Toxoplasma gondii apical end with ICMAP2 conditionally knocked down

EMDB-42120: 
Toxoplasma gondii apical end with ICMAP3i conditionally knocked down

EMDB-42121: 
Toxoplasma gondii apical end with ICMAP3ii conditionally knocked down

EMDB-42527: 
Pre-fusion Measles virus fusion protein complexed with Fab 77

EMDB-42539: 
Structure of the Measles virus Fusion protein in the post-fusion conformation

EMDB-42593: 
Structure of the Measles virus Fusion protein in the pre-fusion conformation

EMDB-42595: 
Structure of the Measles virus Fusion protein in the pre-fusion conformation with bound [FIP-HRC]2-PEG11

EMDB-43827: 
Fab 77-stabilized MeV F ectodomain fragment

PDB-8ut2: 
Pre-fusion Measles virus fusion protein complexed with Fab 77

PDB-8utf: 
Structure of the Measles virus Fusion protein in the post-fusion conformation

PDB-8uup: 
Structure of the Measles virus Fusion protein in the pre-fusion conformation

PDB-8uuq: 
Structure of the Measles virus Fusion protein in the pre-fusion conformation with bound [FIP-HRC]2-PEG11

PDB-9at8: 
Fab 77-stabilized MeV F ectodomain fragment

EMDB-16512: 
MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle

EMDB-16522: 
Structure of ADDoCoV-ADAH11

PDB-8c9n: 
MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle

EMDB-29396: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP

EMDB-29836: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer

EMDB-29880: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)

EMDB-29881: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)

EMDB-29882: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3)

EMDB-29905: 
vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer

PDB-8fr6: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP

PDB-8g85: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer

PDB-8g9w: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)

PDB-8g9x: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)

PDB-8g9y: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3)

PDB-8gas: 
vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer

EMDB-28726: 
Rat 80S ribosome purified from brain RNA granules. Class 1 40S subunit 2_5A resolution

EMDB-28727: 
Rat 80S ribosome purified from brain RNA granules. Class 1 60S subunit 2_5A resolution.

EMDB-26517: 
Rat 80S ribosome purified from brain RNA granules. Class 2 40S subunit.

EMDB-26518: 
Rat 80S ribosome purified from brain RNA granules. Class 2 60S subunit.

EMDB-29538: 
Rat 80S ribosome purified from brain RNA granules. Class 1 80S consensus

EMDB-29539: 
Rat 80S ribosome purified from brain RNA granules. Class 2 80S consensus

EMDB-29044: 
Structure of Zanidatamab bound to HER2

PDB-8ffj: 
Structure of Zanidatamab bound to HER2

EMDB-28558: 
SARS-CoV-2 BA.1 spike ectodomain trimer in complex with the S2X324 neutralizing antibody Fab fragment (local refinement of the RBD and S2X324)

EMDB-28559: 
SARS-CoV-2 Omicron BA.1 spike ectodomain trimer in complex with the S2X324 neutralizing antibody Fab fragment

PDB-8erq: 
SARS-CoV-2 BA.1 spike ectodomain trimer in complex with the S2X324 neutralizing antibody Fab fragment (local refinement of the RBD and S2X324)

PDB-8err: 
SARS-CoV-2 Omicron BA.1 spike ectodomain trimer in complex with the S2X324 neutralizing antibody Fab fragment

EMDB-24599: 
Tomogram of SARS-CoV-2 spike-bearing virus-like particles (VLPs) interacting with hACE2-bearing extracellular vesicles (tEVs), showing various intermediate states of the SARS-CoV-2 spike protein (Fig. 2M,N, Fig 3J, and Supp. Movie 1 of the manuscript Marcink et al., 2021).
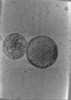
EMDB-24600: 
Tomogram of SARS-CoV-2 spike-bearing virus-like particles (VLPs) interacting with hACE2-bearing extracellular vesicles (tEVs), showing various intermediate states of the SARS-CoV-2 spike protein (Fig. 2K,L, and Supp. Movie 2 of the manuscript Marcink et. al 2021,).

EMDB-24601: 
Tomogram of SARS-CoV-2 spike-bearing virus-like particles (VLPs) interacting with hACE2-bearing extracellular vesicles (tEVs), showing various intermediate states of the SARS-CoV-2 spike protein (Fig. 4B-E and Supp. Movie 3 of manuscript Marcink et al., 2021).
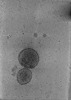
EMDB-24602: 
Tomogram of SARS-CoV-2 spike-bearing virus-like particles (VLPs) interacting with hACE2-bearing extracellular vesicles (tEVs), showing various intermediate states of the SARS-CoV-2 spike protein (Fig 2E-G of the manuscript Marcink et al., 2021).

EMDB-24603: 
Tomogram of SARS-CoV-2 spike-bearing virus-like particles (VLPs) interacting with hACE2-bearing extracellular vesicles (tEVs), showing various intermediate states of the SARS-CoV-2 spike protein (Fig. 3D,E of the manuscript Marcink et al., 2021).
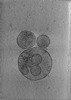
EMDB-24604: 
Tomogram of SARS-CoV-2 spike-bearing virus-like particles (VLPs) interacting with hACE2-bearing extracellular vesicles (tEVs), showing various intermediate states of the SARS-CoV-2 spike protein (Fig. 4H-J of the manuscript Marcink et al., 2021)

EMDB-26679: 
Subtomogram averaged map of hACE2 dimers on the surface of extracellular vesicles

EMDB-25824: 
aRML prion fibril
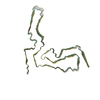
PDB-7td6: 
aRML prion fibril
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
