[English] 日本語
 Yorodumi
Yorodumi- EMDB-42118: Toxoplasma gondii apical end with ICMAP1 conditionally knocked down -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
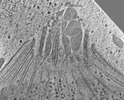
| Title | Toxoplasma gondii apical end with ICMAP1 conditionally knocked down | |||||||||||||||
 Map data Map data | Toxoplasma gondii apical end with ICMAP1 conditionally knocked down | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Parasite / Conoid / Microtubules / CELL INVASION | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | electron tomography / cryo EM | |||||||||||||||
 Authors Authors | Martinez M / Dos Santos Pacheco N / Chang Y-W / Soldati-Favre D | |||||||||||||||
| Funding support |  United States, United States,  Switzerland, European Union, 4 items Switzerland, European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Sustained rhoptry docking and discharge requires Toxoplasma gondii intraconoidal microtubule-associated proteins. Authors: Nicolas Dos Santos Pacheco / Albert Tell I Puig / Amandine Guérin / Matthew Martinez / Bohumil Maco / Nicolò Tosetti / Estefanía Delgado-Betancourt / Matteo Lunghi / Boris Striepen / Yi- ...Authors: Nicolas Dos Santos Pacheco / Albert Tell I Puig / Amandine Guérin / Matthew Martinez / Bohumil Maco / Nicolò Tosetti / Estefanía Delgado-Betancourt / Matteo Lunghi / Boris Striepen / Yi-Wei Chang / Dominique Soldati-Favre /   Abstract: In Apicomplexa, rhoptry discharge is essential for invasion and involves an apical vesicle (AV) docking one or two rhoptries to a macromolecular secretory apparatus. Toxoplasma gondii is armed with ...In Apicomplexa, rhoptry discharge is essential for invasion and involves an apical vesicle (AV) docking one or two rhoptries to a macromolecular secretory apparatus. Toxoplasma gondii is armed with 10-12 rhoptries and 5-6 microtubule-associated vesicles (MVs) presumably for iterative rhoptry discharge. Here, we have addressed the localization and functional significance of two intraconoidal microtubule (ICMT)-associated proteins instrumental for invasion. Mechanistically, depletion of ICMAP2 leads to a dissociation of the ICMTs, their detachment from the conoid and dispersion of MVs and rhoptries. ICMAP3 exists in two isoforms that contribute to the control of the ICMTs length and the docking of the two rhoptries at the AV, respectively. This study illuminates the central role ICMTs play in scaffolding the discharge of multiple rhoptries. This process is instrumental for virulence in the mouse model of infection and in addition promotes sterile protection against T. gondii via the release of key effectors inducing immunity. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42118.map.gz emd_42118.map.gz | 1.2 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42118-v30.xml emd-42118-v30.xml emd-42118.xml emd-42118.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42118.png emd_42118.png | 170.5 KB | ||
| Filedesc metadata |  emd-42118.cif.gz emd-42118.cif.gz | 4.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42118 http://ftp.pdbj.org/pub/emdb/structures/EMD-42118 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42118 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42118 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42118.map.gz / Format: CCP4 / Size: 2.2 GB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) Download / File: emd_42118.map.gz / Format: CCP4 / Size: 2.2 GB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Toxoplasma gondii apical end with ICMAP1 conditionally knocked down | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 10.6 Å | ||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Toxoplasma Gondii tachyzoite apical complex with ICMAP1 condition...
| Entire | Name: Toxoplasma Gondii tachyzoite apical complex with ICMAP1 conditionally knocked down |
|---|---|
| Components |
|
-Supramolecule #1: Toxoplasma Gondii tachyzoite apical complex with ICMAP1 condition...
| Supramolecule | Name: Toxoplasma Gondii tachyzoite apical complex with ICMAP1 conditionally knocked down type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: DMEM supplemented with 5% fetal bovine serum and 2 mM glutamine |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 95 % / Chamber temperature: 310 K / Instrument: LEICA EM GP / Details: Front blot for 4 seconds before plunging. |
| Details | Egressed tachyzoites from a culture of confluent HFF cells. ICMAP1 knockdown induced by treatment with 50 nM rapamycin for 48 hours prior to egress. |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: Ted Pella, Inc / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Software | Name: SerialEM (ver. 3.8) |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 0.4 sec. / Average electron dose: 2.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 33000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Raw frames were gain normalized in serialEM and motion corrected using the alignframes command in IMOD |
|---|---|
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name:  IMOD (ver. 4.11.5) / Number images used: 61 IMOD (ver. 4.11.5) / Number images used: 61 |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)
















