-Search query
-Search result
Showing all 47 items for (author: seifer & s)

EMDB-19762: 
4D_STEM cryo tomogram of t4 Phages produced by PCA2

EMDB-19764: 
4D_STEM cryo tomogram of t4 Phages produced by PCA3

EMDB-19765: 
4D STEM cryo tomogram of t4 Phages produced by iCOM

EMDB-35163: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 5.5

EMDB-35164: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state
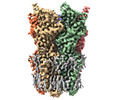
EMDB-36339: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 2.5
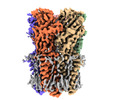
EMDB-37446: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in intermediate state
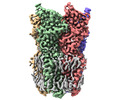
EMDB-37447: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state

PDB-8i47: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 5.5

PDB-8i48: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state

PDB-8jj3: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 2.5

PDB-8wcq: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in intermediate state

PDB-8wcr: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state

EMDB-35161: 
Cryo-EM structure of nanodisc (asolectin) reconstituted GLIC at pH 7.5

EMDB-35162: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 7.5

PDB-8i41: 
Cryo-EM structure of nanodisc (asolectin) reconstituted GLIC at pH 7.5

PDB-8i42: 
Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 7.5

EMDB-19763: 
4D_STEM cryo tomogram of t4 Phages produced by iDPC2

EMDB-16805: 
Cryo-EM structure of PcrV/Fab(30-B8)

EMDB-16807: 
Cryo-EM structure of PcrV/Fab(11-E5)

PDB-8cr9: 
Cryo-EM structure of PcrV/Fab(30-B8)

PDB-8crb: 
Cryo-EM structure of PcrV/Fab(11-E5)

EMDB-27031: 
Accurate computational design of genetically encoded 3D protein crystals

EMDB-40926: 
CryoEM Structure of Computationally Designed Nanocage O32-ZL4

PDB-8cwy: 
Accurate computational design of genetically encoded 3D protein crystals

PDB-8szz: 
CryoEM Structure of Computationally Designed Nanocage O32-ZL4

EMDB-16713: 
Local refinement map of TFIIIC TauB-DNA monomer
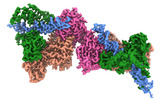
EMDB-16714: 
TFIIIC TauB-DNA dimer
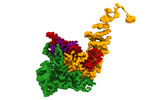
EMDB-16715: 
TFIIIC TauA complex map
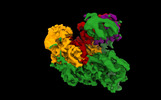
EMDB-16716: 
TFIIIC TauA complex map (sample without DNA)
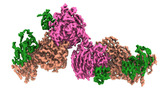
EMDB-16717: 
Structural insights into human TFIIIC promoter recognition

EMDB-17446: 
Consensus map of TauB-DNA dimer (Nu-refinement)

EMDB-17447: 
Local refinement map of TFIIIC TauB-DNA monomer 2

PDB-8cli: 
TFIIIC TauB-DNA monomer

PDB-8clj: 
TFIIIC TauB-DNA dimer

PDB-8clk: 
TFIIIC TauA complex

PDB-8cll: 
Structural insights into human TFIIIC promoter recognition

EMDB-29019: 
Alpha1/BetaB Heteromeric Glycine Receptor in 1 mM Glycine 20 uM Ivermectin State

PDB-8fe1: 
Alpha1/BetaB Heteromeric Glycine Receptor in 1 mM Glycine 20 uM Ivermectin State

EMDB-26130: 
Alpha1/BetaB Heteromeric Glycine Receptor in Strychnine-Bound State
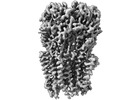
EMDB-26141: 
Alpha1/BetaB Heteromeric Glycine Receptor in Glycine-Bound State

EMDB-15362: 
Cryo-EM structure of Darobactin 22 bound BAM complex

EMDB-15363: 
Cryo-EM structure of Darobactin 9 bound BAM complex

PDB-8adg: 
Cryo-EM structure of Darobactin 22 bound BAM complex

PDB-8adi: 
Cryo-EM structure of Darobactin 9 bound BAM complex

EMDB-11860: 
Subtomogram average structure of anammoxosomal nitrite oxidoreductase

EMDB-11861: 
Helical reconstruction of nitrite oxidoreductase from anammox bacteria
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
