[English] 日本語
 Yorodumi
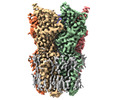
Yorodumi- EMDB-36339: Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 2.5 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 2.5 | |||||||||
 Map data Map data | Postprocess masked map, pH 2.5, GLIC. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | pentameric ligand-gated ion channels / cis-loop / cryo-EM / nanodisc / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsodium channel activity / potassium channel activity / extracellular ligand-gated monoatomic ion channel activity / transmembrane signaling receptor activity / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Gloeobacter violaceus (bacteria) Gloeobacter violaceus (bacteria) | |||||||||
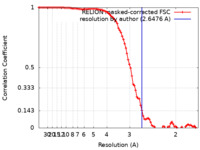
| Method | single particle reconstruction / cryo EM / Resolution: 2.6476 Å | |||||||||
 Authors Authors | Bharambe N / Li Z / Basak S | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment. Authors: Nikhil Bharambe / Zhuowen Li / David Seiferth / Asha Manikkoth Balakrishna / Philip C Biggin / Sandip Basak /   Abstract: GLIC, a proton-activated prokaryotic ligand-gated ion channel, served as a model system for understanding the eukaryotic counterparts due to their structural and functional similarities. Despite ...GLIC, a proton-activated prokaryotic ligand-gated ion channel, served as a model system for understanding the eukaryotic counterparts due to their structural and functional similarities. Despite extensive studies conducted on GLIC, the molecular mechanism of channel gating in the lipid environment requires further investigation. Here, we present the cryo-EM structures of nanodisc-reconstituted GLIC at neutral and acidic pH in the resolution range of 2.6 - 3.4 Å. In our apo state at pH 7.5, the extracellular domain (ECD) displays conformational variations compared to the existing apo structures. At pH 4.0, three distinct conformational states (C1, C2 and O states) are identified. The protonated structures exhibit a compacted and counter-clockwise rotated ECD compared with our apo state. A gradual widening of the pore in the TMD is observed upon reducing the pH, with the widest pore in O state, accompanied by several layers of water pentagons. The pore radius and molecular dynamics (MD) simulations suggest that the O state represents an open conductive state. We also observe state-dependent interactions between several lipids and proteins that may be involved in the regulation of channel gating. Our results provide comprehensive insights into the importance of lipids impact on gating. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36339.map.gz emd_36339.map.gz | 8.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36339-v30.xml emd-36339-v30.xml emd-36339.xml emd-36339.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36339_fsc.xml emd_36339_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_36339.png emd_36339.png | 118.5 KB | ||
| Masks |  emd_36339_msk_1.map emd_36339_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36339.cif.gz emd-36339.cif.gz | 6.4 KB | ||
| Others |  emd_36339_additional_1.map.gz emd_36339_additional_1.map.gz emd_36339_additional_2.map.gz emd_36339_additional_2.map.gz emd_36339_half_map_1.map.gz emd_36339_half_map_1.map.gz emd_36339_half_map_2.map.gz emd_36339_half_map_2.map.gz | 49.4 MB 59.7 MB 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36339 http://ftp.pdbj.org/pub/emdb/structures/EMD-36339 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36339 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36339 | HTTPS FTP |
-Related structure data
| Related structure data |  8jj3MC  8i41C  8i42C  8i47C  8i48C  8wcqC  8wcrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36339.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36339.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocess masked map, pH 2.5, GLIC. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8584 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36339_msk_1.map emd_36339_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
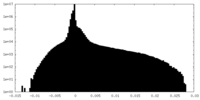
| Density Histograms |
-Additional map: Refinement map, pH 2.5, GLIC.
| File | emd_36339_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement map, pH 2.5, GLIC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
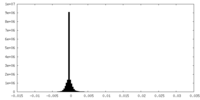
| Density Histograms |
-Additional map: Postprocess unmasked map, pH 2.5, GLIC.
| File | emd_36339_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocess unmasked map, pH 2.5, GLIC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
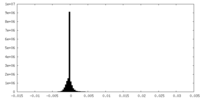
| Density Histograms |
-Half map: Refinement half map 1, pH 2.5, GLIC.
| File | emd_36339_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement half map 1, pH 2.5, GLIC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Refinement half map 2, pH 2.5, GLIC.
| File | emd_36339_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement half map 2, pH 2.5, GLIC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : pentameric ligand-gated ion channel
| Entire | Name: pentameric ligand-gated ion channel |
|---|---|
| Components |
|
-Supramolecule #1: pentameric ligand-gated ion channel
| Supramolecule | Name: pentameric ligand-gated ion channel / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Gloeobacter violaceus (bacteria) Gloeobacter violaceus (bacteria) |
-Macromolecule #1: Proton-gated ion channel
| Macromolecule | Name: Proton-gated ion channel / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Gloeobacter violaceus (bacteria) Gloeobacter violaceus (bacteria) |
| Molecular weight | Theoretical: 35.699094 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VSPPPPIADE PLTVNTGIYL IECYSLDDKA ETFKVNAFLS LSWKDRRLAF DPVRSGVRVK TYEPEAIWIP EIRFVNVENA RDADVVDIS VSPDGTVQYL ERFSARVLSP LDFRRYPFDS QTLHIYLIVR SVDTRNIVLA VDLEKVGKND DVFLTGWDIE S FTAVVKPA ...String: VSPPPPIADE PLTVNTGIYL IECYSLDDKA ETFKVNAFLS LSWKDRRLAF DPVRSGVRVK TYEPEAIWIP EIRFVNVENA RDADVVDIS VSPDGTVQYL ERFSARVLSP LDFRRYPFDS QTLHIYLIVR SVDTRNIVLA VDLEKVGKND DVFLTGWDIE S FTAVVKPA NFALEDRLES KLDYQLRISR QYFSYIPNII LPMLFILFIS WTAFWSTSYE ANVTLVVSTL IAHIAFNILV ET NLPKTPY MTYTGAIIFM IYLFYFVAVI EVTVQHYLKV ESQPARAASI TRASRIAFPV VFLLANIILA FLFFG UniProtKB: Proton-gated ion channel |
-Macromolecule #2: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 2 / Number of copies: 5 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #3: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
| Macromolecule | Name: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine / type: ligand / ID: 3 / Number of copies: 45 / Formula: PEE |
|---|---|
| Molecular weight | Theoretical: 744.034 Da |
| Chemical component information |  ChemComp-PEE: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 265 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 2.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE / Support film - topology: CONTINUOUS | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 6449 / Average exposure time: 6.0 sec. / Average electron dose: 74.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)