-Search query
-Search result
Showing 1 - 50 of 53 items for (author: righetto & r)

EMDB-41105: 
CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3

PDB-8t9a: 
CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3
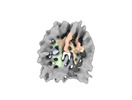
EMDB-19906: 
In situ nucleosome subtomogram average from Chlamydomonas reinhardtii

EMDB-16451: 
Subtomogram average of the T. kivui 70S ribosome in situ

EMDB-15252: 
In situ subtomogram average of the C. reinhardtii stellate at the ciliary transition zone

EMDB-15253: 
In situ subtomogram average of the C. reinhardtii Y-link at the ciliary transition zone

EMDB-15254: 
In situ subtomogram average of the C. reinhardtii MTD sleeve at the ciliary transition zone

EMDB-15255: 
In situ subtomogram average of the C. reinhardtii MTD at the ciliary transition zone

EMDB-15256: 
Composite map of the C. reinhardtii ciliary transition zone (structures attached to a single MTD) from in situ subtomogram averaging

EMDB-15257: 
Composite map of the C. reinhardtii ciliary transition zone (full 9-fold assembly) from in situ subtomogram averaging

EMDB-15258: 
In situ subtomogram average of C. reinhardtii IFT-B at the ciliary transition zone

EMDB-15259: 
In situ subtomogram average of C. reinhardtii IFT-A at the ciliary transition zone

EMDB-15260: 
In situ subtomogram average of C. reinhardtii dynein-1b at the ciliary transition zone

EMDB-15261: 
Composite map of the C. reinhardtii IFT (segment of a fully assembled train) at the ciliary transition zone

EMDB-15262: 
Tomogram #3 of the C. reinhardtii ciliary transition zone

EMDB-15263: 
Tomogram #12 of the C. reinhardtii ciliary transition zone
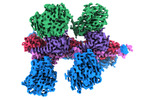
EMDB-14169: 
Cryo-EM structure of Hydrogen-dependent CO2 reductase.

EMDB-15053: 
In situ structure of HDCR filaments

EMDB-15054: 
In situ structure of the T. kivui ribosome

EMDB-15055: 
In situ cryo-electron tomogram of a T. kivui cell 1

EMDB-15056: 
In situ cryo-electron tomogram of a T. kivui cell 2

PDB-7qv7: 
Cryo-EM structure of Hydrogen-dependent CO2 reductase.

EMDB-12073: 
Cryo-EM Structure of Human Thyroglobulin

PDB-7b75: 
Cryo-EM Structure of Human Thyroglobulin

EMDB-10615: 
FAK structure from single particle analysis of 2D crystals

EMDB-10616: 
FAK structure with AMP-PNP from single particle analysis of 2D crystals

PDB-6ty3: 
FAK structure from single particle analysis of 2D crystals

PDB-6ty4: 
FAK structure with AMP-PNP from single particle analysis of 2D crystals

EMDB-10835: 
High resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica

PDB-6yl3: 
High resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica

EMDB-4628: 
Cryo-EM structure of Tobacco Mossaic Virus from microfluidic grid preparation.

EMDB-4738: 
Endogeneous native human 20S proteasome with bound Fabs isolated from less than 1 ul cell lysate

PDB-6r70: 
Endogeneous native human 20S proteasome

PDB-6r7m: 
Tobacco Mosaic Virus (TMV)

EMDB-4432: 
MloK1 consensus map from single particle analysis of 2D crystals

EMDB-4439: 
Full MloK1 consensus map from single particle analysis of 2D crystals

EMDB-4441: 
MloK1 map from single particle analysis of 2D crystals, class 1 (extended conformation)

EMDB-4513: 
MloK1 map from single particle analysis of 2D crystals, class 2 (intermediate conformation)

EMDB-4514: 
MloK1 map from single particle analysis of 2D crystals, class 3 (intermediate extended conformation)

EMDB-4515: 
MloK1 map from single particle analysis of 2D crystals, class 4 (compact/open conformation)

EMDB-4516: 
MloK1 map from single particle analysis of 2D crystals, class 5 (intermediate compact conformation)

EMDB-4517: 
MloK1 map from single particle analysis of 2D crystals, class 6 (intermediate compact conformation)

EMDB-4518: 
MloK1 map from single particle analysis of 2D crystals, class 7 (intermediate conformation)

EMDB-4519: 
MloK1 model from single particle analysis of 2D crystals, class 8 (intermediate conformation)

PDB-6i9d: 
MloK1 consensus structure from single particle analysis of 2D crystals

PDB-6iax: 
MloK1 model from single particle analysis of 2D crystals, class 1 (extended conformation)

PDB-6qcy: 
MloK1 model from single particle analysis of 2D crystals, class 2 (intermediate conformation)

PDB-6qcz: 
MloK1 model from single particle analysis of 2D crystals, class 3 (intermediate extended conformation)

PDB-6qd0: 
MloK1 model from single particle analysis of 2D crystals, class 4 (compact/open conformation)

PDB-6qd1: 
MloK1 model from single particle analysis of 2D crystals, class 5 (intermediate compact conformation)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
