[English] 日本語
 Yorodumi
Yorodumi- EMDB-4439: Full MloK1 consensus map from single particle analysis of 2D crystals -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4439 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Full MloK1 consensus map from single particle analysis of 2D crystals | |||||||||
 Map data Map data | Full MloK1 consensus map from single particle analysis of 2D crystals comprising 2x2 unit cells, sharpened. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Mesorhizobium loti MAFF303099 (bacteria) Mesorhizobium loti MAFF303099 (bacteria) | |||||||||
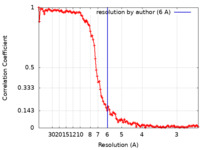
| Method | single particle reconstruction / cryo EM / Resolution: 6.0 Å | |||||||||
 Authors Authors | Righetto R / Biyani N / Kowal J / Chami M / Stahlberg H | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM. Authors: Ricardo D Righetto / Nikhil Biyani / Julia Kowal / Mohamed Chami / Henning Stahlberg /  Abstract: Electron crystallography can reveal the structure of membrane proteins within 2D crystals under close-to-native conditions. High-resolution structural information can only be reached if crystals are ...Electron crystallography can reveal the structure of membrane proteins within 2D crystals under close-to-native conditions. High-resolution structural information can only be reached if crystals are perfectly flat and highly ordered. In practice, such crystals are difficult to obtain. Available image unbending algorithms correct for disorder, but only perform well on images of non-tilted, flat crystals, while out-of-plane distortions are not addressed. Here, we present an approach that employs single-particle refinement procedures to locally unbend crystals in 3D. With this method, density maps of the MloK1 potassium channel with a resolution of 4 Å were obtained from images of 2D crystals that do not diffract beyond 10 Å. Furthermore, 3D classification allowed multiple structures to be resolved, revealing a series of MloK1 conformations within a single 2D crystal. This conformational heterogeneity explains the poor diffraction observed and is related to channel function. The approach is implemented in the FOCUS package. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4439.map.gz emd_4439.map.gz | 116.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4439-v30.xml emd-4439-v30.xml emd-4439.xml emd-4439.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4439_fsc.xml emd_4439_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_4439.png emd_4439.png | 68.7 KB | ||
| Others |  emd_4439_half_map_1.map.gz emd_4439_half_map_1.map.gz emd_4439_half_map_2.map.gz emd_4439_half_map_2.map.gz | 47.5 MB 47.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4439 http://ftp.pdbj.org/pub/emdb/structures/EMD-4439 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4439 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4439 | HTTPS FTP |
-Related structure data
| Related structure data |  4432C  4441C  4513C  4514C  4515C  4516C  4517C  4518C  4519C  6i9dC  6iaxC  6qcyC  6qczC  6qd0C  6qd1C  6qd2C  6qd3C  6qd4C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4439.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4439.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full MloK1 consensus map from single particle analysis of 2D crystals comprising 2x2 unit cells, sharpened. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_4439_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_4439_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MloK1 potassium channel in lipidic bilayer of 2D crystals
| Entire | Name: MloK1 potassium channel in lipidic bilayer of 2D crystals |
|---|---|
| Components |
|
-Supramolecule #1: MloK1 potassium channel in lipidic bilayer of 2D crystals
| Supramolecule | Name: MloK1 potassium channel in lipidic bilayer of 2D crystals type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Mesorhizobium loti MAFF303099 (bacteria) Mesorhizobium loti MAFF303099 (bacteria) |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 160 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 Details: 20 mM KCl, 20 mM Tris-HCl pH 7.6, 1 mM BaCl2, 1 mM EDTA, 0.2 mM cAMP |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 270 / Average exposure time: 16.0 sec. / Average electron dose: 45.0 e/Å2 Details: The dataset of 346 movies recorded and processed for Kowal et al. (2018) 2 was employed. As reported, the data were collected on an FEI Titan Krios TEM equipped with a Gatan K2 DED. Total ...Details: The dataset of 346 movies recorded and processed for Kowal et al. (2018) 2 was employed. As reported, the data were collected on an FEI Titan Krios TEM equipped with a Gatan K2 DED. Total dose: 40 e-/A^2 distributed over 40 movie frames. Pixel size: 1.3 A on the sample level (counting mode). Nominal tilt range: -55 to +55 degrees. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.3 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)