-Search query
-Search result
Showing all 38 items for (author: pinke & g)

EMDB-17929: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: pooled class

EMDB-17930: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -20 degrees

EMDB-17931: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -15 degrees

EMDB-17932: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -10 degrees

EMDB-17933: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -05 degrees

EMDB-17934: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 0 degrees

EMDB-17935: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 05 degrees

EMDB-17936: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 10 degrees

EMDB-17937: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 15 degrees

EMDB-17938: 
Cryo-electron tomogram of HTLV-1 MA126CANC tubes

EMDB-17939: 
Cryo-electron tomogram of HTLV-1 MA126CANC tubes

EMDB-17940: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 20 degrees

EMDB-17941: 
Cryo-electron tomogram containing HTLV-1 Gag-based VLPs

EMDB-17942: 
Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-NTD refinement

EMDB-17943: 
Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-CTD refinement

PDB-8pu6: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: pooled class

PDB-8pu7: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -20 degrees

PDB-8pu8: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -15 degrees

PDB-8pu9: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -10 degrees

PDB-8pua: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -05 degrees

PDB-8pub: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 0 degrees

PDB-8puc: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 05 degrees

PDB-8pud: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 10 degrees

PDB-8pue: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 15 degrees

PDB-8puf: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 20 degrees

PDB-8pug: 
Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-NTD refinement

PDB-8puh: 
Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-CTD refinement
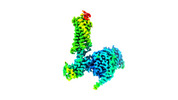
EMDB-29301: 
Neurotensin receptor allosterism revealed in complex with a biased allosteric modulator

EMDB-29302: 
CryoEM structure of Go-coupled NTSR1 with a biased allosteric modulator
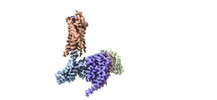
EMDB-29303: 
CryoEM structure of Go-coupled NTSR1

PDB-8fmz: 
Neurotensin receptor allosterism revealed in complex with a biased allosteric modulator

PDB-8fn0: 
CryoEM structure of Go-coupled NTSR1 with a biased allosteric modulator

PDB-8fn1: 
CryoEM structure of Go-coupled NTSR1

EMDB-26987: 
T148-ADPR-F-actin

EMDB-10573: 
Ovine ATP synthase 1a state

EMDB-11127: 
Fo domain of Ovine ATP synthase

PDB-6tt7: 
Ovine ATP synthase 1a state

PDB-6za9: 
Fo domain of Ovine ATP synthase
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
