-Search query
-Search result
Showing 1 - 50 of 352 items for (author: min & ds)

EMDB-40825: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10

PDB-8sx3: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10

EMDB-40815: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, N611 and base from participant 017

EMDB-40816: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp41-N611/FP and base from participant 03

EMDB-40817: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp41-N611/FP and base from participant 07

EMDB-40818: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies gp120-GH and base from participant 09

EMDB-40819: 
BG505 SOSIP.664 in complex with wk26 human polyclonal antibodies C3V5, V1V3, gp41-GH/FP and base from participant 11

EMDB-41363: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF5

PDB-8tl6: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF5

EMDB-40603: 
GPR161 Gs heterotrimer
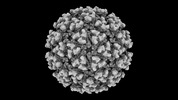
EMDB-42187: 
Eastern equine encephalitis virus (PE-6) VLP (icosahedral)

EMDB-42189: 
Eastern equine encephalitis virus (PE-6) VLP in complex with full-length VLDLR (icosahedral)
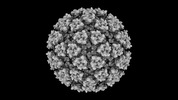
EMDB-42191: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-2) (icosahedral)

EMDB-42193: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-3) (icosahedral)
EMDB-42194: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-3) (asymmetric unit)

EMDB-42195: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-5)[W132A] (icosahedral)

EMDB-42196: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-5)[W132A] (asymmetric unit)
EMDB-42197: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W132A, W210A] (icosahedral)

EMDB-42198: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W132A, W210A] (asymmetric unit)
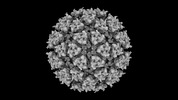
EMDB-42199: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-8)[W132A, W210A, W256A] (icosahedral)

EMDB-42200: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-8)[W132A, W210A, W256A] (asymmetric unit)
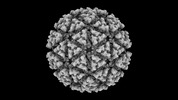
EMDB-42201: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(3-8) (icosahedral)

EMDB-42202: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W89A] (icosahedral)

EMDB-42203: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-6)[W89A] (asymmetric unit)
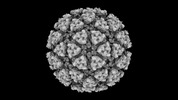
EMDB-42212: 
Eastern equine encephalitis virus (FL93-939) VLP in complex with VLDLR LA(1-6) (icosahedral)

EMDB-42188: 
Eastern equine encephalitis virus (PE-6) VLP (asymmetric unit)

EMDB-42190: 
Eastern equine encephalitis virus (PE-6) VLP in complex with full-length VLDLR (asymmetric unit)

EMDB-42192: 
Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-2) (asymmetric unit)
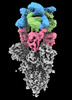
EMDB-16453: 
SARS-CoV-2 Omicron Variant Spike Trimer in complex with three 17T2 Fabs

EMDB-16473: 
SARS-CoV-2 spike in complex with the 17T2 neutralizing antibody Fab fragment (local refinement of RBD and Fab)

PDB-8c89: 
SARS-CoV-2 spike in complex with the 17T2 neutralizing antibody Fab fragment (local refinement of RBD and Fab)

EMDB-41174: 
ssRNA bound SAMHD1 T closed

PDB-8tdv: 
ssRNA bound SAMHD1 T closed

EMDB-41171: 
Cryo-EM structure of cardiac amyloid fibril from a variant ATTR I84S amyloidosis patient-3, wild-type morphology

EMDB-41172: 
Cryo-EM structure of cardiac amyloid fibril from a variant ATTR I84S amyloidosis patient-3, variant-type morphology

EMDB-41788: 
S. cerevisiae Pex1/Pex6 with 1 mM ATP

PDB-8u0v: 
S. cerevisiae Pex1/Pex6 with 1 mM ATP

EMDB-41175: 
ssRNA bound SAMHD1 T open

PDB-8tdw: 
ssRNA bound SAMHD1 T open
EMDB-15985: 
Small molecule positive allosteric modulation of homomeric kainate receptors GluK1-3: Development of screening assays and insight into GluK3 structure

EMDB-15986: 
Ionotropic glutamate receptor K3 with glutamate and BPAM

EMDB-28036: 
Cryo-EM structure of the full-length human NF1 dimer

PDB-8edm: 
Cryo-EM structure of the full-length human NF1 dimer

EMDB-41158: 
CS2it1p2_F7K local refinement for HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab

EMDB-41180: 
Global reconstruction for HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab

EMDB-16963: 
Leishmania tarentolae proteasome 20S subunit in complex with 1-Benzyl-N-(3-(cyclopropylcarbamoyl)phenyl)-6-oxo-1,6-dihydropyridazine-3-carboxamide

PDB-8olu: 
Leishmania tarentolae proteasome 20S subunit in complex with 1-Benzyl-N-(3-(cyclopropylcarbamoyl)phenyl)-6-oxo-1,6-dihydropyridazine-3-carboxamide

EMDB-17756: 
Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein

PDB-8pm2: 
Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein

EMDB-27703: 
Structure of RBD directed antibody DH1047 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interace
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
