[English] 日本語
 Yorodumi
Yorodumi- EMDB-42199: Eastern equine encephalitis virus (PE-6) VLP in complex with VLDL... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Eastern equine encephalitis virus (PE-6) VLP in complex with VLDLR LA(1-8)[W132A, W210A, W256A] (icosahedral) | |||||||||
 Map data Map data | EEEV PE-6 VLP in complex with VLDLR LA(1-8) W132A, W210A, W256A icosahedral reconstruction | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | virus / receptor / VIRUS LIKE PARTICLE | |||||||||
| Biological species |   Eastern equine encephalitis virus Eastern equine encephalitis virus | |||||||||
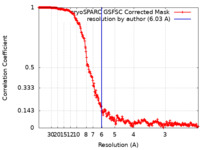
| Method | single particle reconstruction / cryo EM / Resolution: 6.03 Å | |||||||||
 Authors Authors | Adams LJ / Fremont DH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Structural and functional basis of VLDLR usage by Eastern equine encephalitis virus. Authors: Lucas J Adams / Saravanan Raju / Hongming Ma / Theron Gilliland / Douglas S Reed / William B Klimstra / Daved H Fremont / Michael S Diamond /  Abstract: The very-low-density lipoprotein receptor (VLDLR) comprises eight LDLR type A (LA) domains and supports entry of distantly related alphaviruses, including Eastern equine encephalitis virus (EEEV) and ...The very-low-density lipoprotein receptor (VLDLR) comprises eight LDLR type A (LA) domains and supports entry of distantly related alphaviruses, including Eastern equine encephalitis virus (EEEV) and Semliki Forest virus (SFV). Here, by resolving multiple cryo-electron microscopy structures of EEEV-VLDLR complexes and performing mutagenesis and functional studies, we show that EEEV uses multiple sites (E1/E2 cleft and E2 A domain) to engage more than one LA domain simultaneously. However, no single LA domain is necessary or sufficient to support efficient EEEV infection. Whereas all EEEV strains show conservation of two VLDLR-binding sites, the EEEV PE-6 strain and a few other EEE complex members feature a single amino acid substitution that enables binding of LA domains to an additional site on the E2 B domain. These structural and functional analyses informed the design of a minimal VLDLR decoy receptor that neutralizes EEEV infection and protects mice from lethal challenge. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42199.map.gz emd_42199.map.gz | 1.3 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42199-v30.xml emd-42199-v30.xml emd-42199.xml emd-42199.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42199_fsc.xml emd_42199_fsc.xml | 25.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_42199.png emd_42199.png | 51 KB | ||
| Filedesc metadata |  emd-42199.cif.gz emd-42199.cif.gz | 3.9 KB | ||
| Others |  emd_42199_additional_1.map.gz emd_42199_additional_1.map.gz emd_42199_half_map_1.map.gz emd_42199_half_map_1.map.gz emd_42199_half_map_2.map.gz emd_42199_half_map_2.map.gz | 698.9 MB 1.3 GB 1.3 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42199 http://ftp.pdbj.org/pub/emdb/structures/EMD-42199 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42199 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42199 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42199.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42199.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EEEV PE-6 VLP in complex with VLDLR LA(1-8) W132A, W210A, W256A icosahedral reconstruction | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.184 Å | ||||||||||||||||||||||||||||||||||||
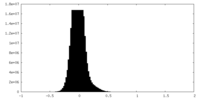
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: EEEV PE-6 VLP in complex with VLDLR LA(1-8)...
| File | emd_42199_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EEEV PE-6 VLP in complex with VLDLR LA(1-8) W132A, W210A, W256A icosahedral reconstruction (unsharpened map) | ||||||||||||
| Projections & Slices |
| ||||||||||||
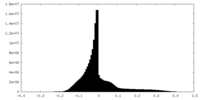
| Density Histograms |
-Half map: EEEV PE-6 VLP in complex with VLDLR LA(1-8)...
| File | emd_42199_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EEEV PE-6 VLP in complex with VLDLR LA(1-8) W132A, W210A, W256A icosahedral reconstruction (half map A) | ||||||||||||
| Projections & Slices |
| ||||||||||||
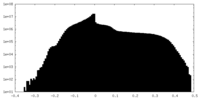
| Density Histograms |
-Half map: EEEV PE-6 VLP in complex with VLDLR LA(1-8)...
| File | emd_42199_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EEEV PE-6 VLP in complex with VLDLR LA(1-8) W132A, W210A, W256A icosahedral reconstruction (half map B) | ||||||||||||
| Projections & Slices |
| ||||||||||||
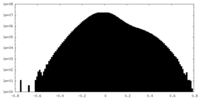
| Density Histograms |
- Sample components
Sample components
-Entire : Eastern equine encephalitis virus
| Entire | Name:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
|---|---|
| Components |
|
-Supramolecule #1: Eastern equine encephalitis virus
| Supramolecule | Name: Eastern equine encephalitis virus / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 11021 / Sci species name: Eastern equine encephalitis virus / Sci species strain: PE-6 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: Yes |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 39.9 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.9000000000000001 µm / Nominal defocus min: 0.5 µm |
 Movie
Movie Controller
Controller


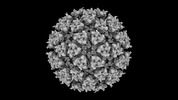




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)