-Search query
-Search result
Showing 1 - 50 of 769 items for (author: li & yc)

EMDB-42291: 
Structure of the human INTS9-INTS11-BRAT1 complex

EMDB-42292: 
Structure of the Drosophila IntS11-CG7044(dBRAT1) complex

EMDB-39212: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH8.0 (3.23A)

EMDB-39213: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH6.5 (2.82A)

EMDB-39214: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH5.0 (3.52A)

EMDB-39215: 
Cryo-EM structure of Dragon Grouper nervous necrosis virion at pH6.5 (3.12A)

EMDB-39217: 
Cryo-EM structure of Dragon Grouper nervous necrosis virion at pH5.0 (4.36A)

EMDB-17696: 
Structure of human 48S translation initiation complex in open codon scanning state (48S-1)

EMDB-17697: 
Structure of human 48S translation initiation complex in AUG recognition state after eIF5-induced GTP hydrolysis by eIF2 (48S-2)

EMDB-17698: 
Structure of human 48S translation initiation complex upon transfer of initiator tRNA to eIF5B (48S-3)

EMDB-17699: 
Structure of human 48S translation initiation complex after eIF5 release (48S-4)

EMDB-17700: 
Structure of human 48S translation initiation complex after eIF2 release prior 60S subunit joining (48S-5)

EMDB-17701: 
Structure of human 48S translation initiation complex with initiator tRNA, eIF1A and eIF3 (off-pathway)

EMDB-19128: 
Structure of human eIF3 core from closed 48S translation initiation complex

PDB-8pj1: 
Structure of human 48S translation initiation complex in open codon scanning state (48S-1)

PDB-8pj2: 
Structure of human 48S translation initiation complex in AUG recognition state after eIF5-induced GTP hydrolysis by eIF2 (48S-2)

PDB-8pj3: 
Structure of human 48S translation initiation complex upon transfer of initiator tRNA to eIF5B (48S-3)

PDB-8pj4: 
Structure of human 48S translation initiation complex after eIF5 release (48S-4)

PDB-8pj5: 
Structure of human 48S translation initiation complex after eIF2 release prior 60S subunit joining (48S-5)

PDB-8pj6: 
Structure of human 48S translation initiation complex with initiator tRNA, eIF1A and eIF3 (off-pathway)

PDB-8rg0: 
Structure of human eIF3 core from closed 48S translation initiation complex

EMDB-43700: 
Cryo-EM map of LKB1-STRADalpha-MO25alpha from TFS Glacios with Gatan Alpine detector at 120 keV

EMDB-43701: 
Cryo-EM map of LKB1-STRADalpha-MO25alpha from TFS Glacios with Gatan Alpine detector at 200 keV

EMDB-43702: 
Cryo-EM map of LKB1-STRADalpha-MO25alpha from TFS Glacios with Gatan K3 detector at 200 keV

EMDB-39025: 
Structure of HCoV-HKU1A spike in the functionally anchored-3up conformation with 3TMPRSS2

EMDB-39036: 
Structure of HCoV-HKU1C spike in the functionally anchored-1up conformation with 1TMPRSS2

EMDB-39037: 
Structure of HCoV-HKU1C spike in the functionally anchored-2up conformation with 2TMPRSS2

EMDB-39038: 
Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 2TMPRSS2

EMDB-39039: 
Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 3TMPRSS2

EMDB-39040: 
Local structure of HCoV-HKU1C spike in complex with TMPRSS2 and glycan

EMDB-39041: 
Structure of HCoV-HKU1C spike in the inactive-closed conformation

EMDB-39042: 
Structure of HCoV-HKU1C spike in the inactive-1up conformation

EMDB-39043: 
Structure of HCoV-HKU1C spike in the inactive-2up conformation

EMDB-39044: 
Structure of HCoV-HKU1C spike in the glycan-activated-closed conformation

EMDB-39045: 
Structure of HCoV-HKU1C spike in the glycan-activated-1up conformation

EMDB-39046: 
Structure of HCoV-HKU1C spike in the glycan-activated-2up conformation

EMDB-39047: 
Structure of HCoV-HKU1C spike in the glycan-activated-3up conformation

EMDB-39048: 
Local structure of HCoV-HKU1C spike in complex with glycan

EMDB-43506: 
Cryo-EM structure of LKB1-STRADalpha-MO25alpha heterocomplex

PDB-8vsu: 
Cryo-EM structure of LKB1-STRADalpha-MO25alpha heterocomplex

EMDB-43527: 
Apoferritin at 100 keV on Alpine detector with a side-entry cryoholder
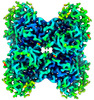
EMDB-43528: 
Aldolase at 100 keV on the Alpine detector with a side-entry cryoholder

EMDB-38216: 
Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2
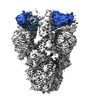
EMDB-39546: 
SARS-CoV-2 Delta Spike in complex with JL-8C

EMDB-39547: 
SARS-CoV-2 Delta Spike in complex with JM-1A

EMDB-39685: 
SARS-CoV-2 Delta Spike in complex with Fab of JE-5C

EMDB-39686: 
SARS-CoV-2 Spike (BA.1) in complex with Fab of JH-8B

EMDB-29330: 
N332-GT5 SOSIP in complex with base polyclonal Fabs isolated at day 42 from protein immunized wild type mice

EMDB-29333: 
N332-GT5 SOSIP in complex with base polyclonal Fabs isolated at day 42 from protein immunized BG18HCgl knock-in mice

EMDB-29334: 
N332-GT5 SOSIP in complex with V1V3 polyclonal Fabs isolated at day 16 from mRNA immunized wild type mice
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
