[English] 日本語
 Yorodumi
Yorodumi- EMDB-19128: Structure of human eIF3 core from closed 48S translation initiati... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human eIF3 core from closed 48S translation initiation complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RIBOSOME / TRANSLATION / initiation / 48S / eIF / human / eukaryotic / factor / codon / scanning / open / reading | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of mRNA binding / viral translational termination-reinitiation / eukaryotic translation initiation factor 3 complex, eIF3e / cap-dependent translational initiation / eukaryotic translation initiation factor 3 complex, eIF3m / IRES-dependent viral translational initiation / translation reinitiation / formation of cytoplasmic translation initiation complex / multi-eIF complex / cytoplasmic translational initiation ...positive regulation of mRNA binding / viral translational termination-reinitiation / eukaryotic translation initiation factor 3 complex, eIF3e / cap-dependent translational initiation / eukaryotic translation initiation factor 3 complex, eIF3m / IRES-dependent viral translational initiation / translation reinitiation / formation of cytoplasmic translation initiation complex / multi-eIF complex / cytoplasmic translational initiation / eukaryotic translation initiation factor 3 complex / eukaryotic 43S preinitiation complex / mRNA cap binding / eukaryotic 48S preinitiation complex / regulation of translational initiation / metal-dependent deubiquitinase activity / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / negative regulation of RNA splicing / rRNA modification in the nucleus and cytosol / erythrocyte homeostasis / Formation of the ternary complex, and subsequently, the 43S complex / cytoplasmic side of rough endoplasmic reticulum membrane / laminin receptor activity / Ribosomal scanning and start codon recognition / Translation initiation complex formation / SARS-CoV-1 modulates host translation machinery / Peptide chain elongation / Selenocysteine synthesis / Formation of a pool of free 40S subunits / Eukaryotic Translation Termination / SRP-dependent cotranslational protein targeting to membrane / Response of EIF2AK4 (GCN2) to amino acid deficiency / Viral mRNA Translation / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / GTP hydrolysis and joining of the 60S ribosomal subunit / L13a-mediated translational silencing of Ceruloplasmin expression / Major pathway of rRNA processing in the nucleolus and cytosol / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / laminin binding / translation regulator activity / translation initiation factor binding / negative regulation of translational initiation / translation initiation factor activity / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / negative regulation of proteasomal ubiquitin-dependent protein catabolic process / antiviral innate immune response / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / cytosolic ribosome / Resolution of Sister Chromatid Cohesion / positive regulation of translation / erythrocyte differentiation / maturation of SSU-rRNA / translational initiation / small-subunit processome / RHO GTPases Activate Formins / PML body / receptor tyrosine kinase binding / negative regulation of ERK1 and ERK2 cascade / mRNA 5'-UTR binding / GABA-ergic synapse / Regulation of expression of SLITs and ROBOs / fibrillar center / metallopeptidase activity / rRNA processing / Separation of Sister Chromatids / presynapse / ribosome biogenesis / ribosome binding / virus receptor activity / ribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / microtubule / SARS-CoV-2 modulates host translation machinery / cytoplasmic translation / cell differentiation / ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity / postsynaptic density / structural constituent of ribosome / cadherin binding / ribosome / translation / ribonucleoprotein complex / focal adhesion / mRNA binding / synapse / negative regulation of apoptotic process / chromatin / nucleolus / glutamatergic synapse / structural molecule activity / negative regulation of transcription by RNA polymerase II / endoplasmic reticulum / proteolysis / DNA binding / RNA binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
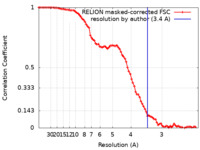
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Petrychenko V / Yi S-H / Liedtke D / Peng BZ / Rodnina MV / Fischer N | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Structural basis for translational control by the human 48S initiation complex. Authors: Valentyn Petrychenko / Sung-Hui Yi / David Liedtke / Bee-Zen Peng / Marina V Rodnina / Niels Fischer /  Abstract: The selection of an open reading frame (ORF) for translation of eukaryotic mRNA relies on remodeling of the scanning 48S initiation complex into an elongation-ready 80S ribosome. Using cryo-electron ...The selection of an open reading frame (ORF) for translation of eukaryotic mRNA relies on remodeling of the scanning 48S initiation complex into an elongation-ready 80S ribosome. Using cryo-electron microscopy, we visualize the key commitment steps orchestrating 48S remodeling in humans. The mRNA Kozak sequence facilitates mRNA scanning in the 48S open state and stabilizes the 48S closed state by organizing the contacts of eukaryotic initiation factors (eIFs) and ribosomal proteins and by reconfiguring mRNA structure. GTPase-triggered large-scale fluctuations of 48S-bound eIF2 facilitate eIF5B recruitment, transfer of initiator tRNA from eIF2 to eIF5B and the release of eIF5 and eIF2. The 48S-bound multisubunit eIF3 complex controls ribosomal subunit joining by coupling eIF exchange to gradual displacement of the eIF3c N-terminal domain from the intersubunit interface. These findings reveal the structural mechanism of ORF selection in human cells and explain how eIF3 could function in the context of the 80S ribosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19128.map.gz emd_19128.map.gz | 47.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19128-v30.xml emd-19128-v30.xml emd-19128.xml emd-19128.xml | 47.9 KB 47.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19128_fsc.xml emd_19128_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_19128.png emd_19128.png | 206.8 KB | ||
| Masks |  emd_19128_msk_1.map emd_19128_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19128.cif.gz emd-19128.cif.gz | 13.3 KB | ||
| Others |  emd_19128_additional_1.map.gz emd_19128_additional_1.map.gz emd_19128_additional_2.map.gz emd_19128_additional_2.map.gz emd_19128_half_map_1.map.gz emd_19128_half_map_1.map.gz emd_19128_half_map_2.map.gz emd_19128_half_map_2.map.gz | 49.4 MB 40.6 MB 40.7 MB 40.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19128 http://ftp.pdbj.org/pub/emdb/structures/EMD-19128 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19128 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19128 | HTTPS FTP |
-Related structure data
| Related structure data |  8rg0MC  8pj1C  8pj2C  8pj3C  8pj4C  8pj5C  8pj6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19128.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19128.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19128_msk_1.map emd_19128_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map from focused flexible refinement with improved density...
| File | emd_19128_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map from focused flexible refinement with improved density for eIF3k/l | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened main map from 3D refinement step
| File | emd_19128_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened main map from 3D refinement step | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_19128_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_19128_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Human 48S initiation complex 40S-eIF1-eIF1A-eIF2-eIF3-tRNA-Met-mRNA
+Supramolecule #1: Human 48S initiation complex 40S-eIF1-eIF1A-eIF2-eIF3-tRNA-Met-mRNA
+Supramolecule #2: Human 48S initiation complex 40S-eIF1-eIF1A-eIF2-eIF3-tRNA-Met
+Supramolecule #3: mRNA
+Macromolecule #1: Eukaryotic translation initiation factor 3 subunit K
+Macromolecule #2: Eukaryotic translation initiation factor 3 subunit F
+Macromolecule #3: Eukaryotic translation initiation factor 3 subunit L
+Macromolecule #4: Eukaryotic translation initiation factor 3 subunit M
+Macromolecule #6: Eukaryotic translation initiation factor 3 subunit H
+Macromolecule #8: 40S ribosomal protein S27
+Macromolecule #9: 40S ribosomal protein S13
+Macromolecule #10: 40S ribosomal protein S17
+Macromolecule #11: 40S ribosomal protein SA
+Macromolecule #12: 40S ribosomal protein S3a
+Macromolecule #13: 40S ribosomal protein S14
+Macromolecule #14: 40S ribosomal protein S26
+Macromolecule #15: 40S ribosomal protein S28
+Macromolecule #16: Eukaryotic translation initiation factor 3 subunit A
+Macromolecule #17: Eukaryotic translation initiation factor 3 subunit E
+Macromolecule #18: Eukaryotic translation initiation factor 3 subunit D
+Macromolecule #19: Eukaryotic translation initiation factor 3 subunit C
+Macromolecule #5: mRNA
+Macromolecule #7: 18S rRNA
+Macromolecule #20: MAGNESIUM ION
+Macromolecule #21: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: HOMEMADE PLUNGER / Details: Manual blotting & plunge-freezing. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Electron-optical aberrations were corrected using a CETCOR Cs-corrector (CEOS, Heidelberg) aligned with the CETCORPLUS 4.6.9 software package (CEOS, Heidelberg). |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average exposure time: 1.5 sec. / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.2 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-8rg0: |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)