-Search query
-Search result
Showing 1 - 50 of 139 items for (author: lee & cw)

EMDB-17377: 
Structure of human SIT1 (focussed map / refinement)

EMDB-17378: 
Structure of human SIT1:ACE2 complex (open PD conformation)

EMDB-17379: 
Structure of human SIT1:ACE2 complex (closed PD conformation)

EMDB-17380: 
Structure of human SIT1 bound to L-pipecolate (focussed map / refinement)

EMDB-17381: 
Structure of human SIT1:ACE2 complex (open PD conformation) bound to L-pipecolate

EMDB-17382: 
Structure of human SIT1:ACE2 complex (closed PD conformation) bound to L-pipecolate

EMDB-35010: 
Human Consensus Olfactory Receptor OR52c in Complex with Octanoic Acid (OCA) and G Protein

EMDB-35770: 
Human Consensus Olfactory Receptor OR52c in Complex with Octanoic Acid (OCA) and G Protein (Consensus map)

EMDB-35772: 
Human Consensus Olfactory Receptor OR52c in Complex with Octanoic Acid (OCA) and G Protein (Receptor-focused map)

EMDB-35773: 
Human Consensus Olfactory Receptor OR52c in Complex with Octanoic Acid (OCA) and G Protein (G protein-focused map)
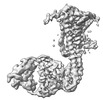
EMDB-35971: 
Human Consensus Olfactory Receptor OR52c in apo state, OR52c-bRIL
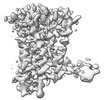
EMDB-37336: 
Human Consensus Olfactory Receptor OR52c in apo state, OR52c only

EMDB-28552: 
Human Membrane-bound O-acyltransferase 7

EMDB-33650: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7

EMDB-33651: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface)

EMDB-33872: 
Human Cytosolic 10-formyltetrahydrofolate dehydrogenase and Gossypol complex

EMDB-28577: 
Human triacylglycerol synthesizing enzyme DGAT1 in complex with T863 inhibitor

EMDB-28594: 
Human triacylglycerol synthesizing enzyme DGAT1 in complex with DGAT1IN1 inhibitor

EMDB-33393: 
Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs (C1 symmetry)

EMDB-33394: 
Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs

EMDB-33392: 
Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in POPE/CHS nanodiscs at pH ~8.0

EMDB-33391: 
Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in GDN detergents at pH ~8.0

EMDB-33395: 
Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (GCN conformation)

EMDB-33396: 
Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (GCN-TM1i conformation)

EMDB-33397: 
Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (FIN conformation)

EMDB-33398: 
Hemichannel-focused structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel in POPE nanodiscs (PLN conformation)

EMDB-33399: 
Consensus map of connexin43/Cx43/GJA1 gap junction intercellular channel in LMNG/CHS detergents at pH ~6.9
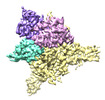
EMDB-27898: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA
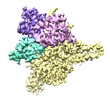
EMDB-27899: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA

EMDB-15526: 
In situ cryo-electron tomogram of a bulk autophagy phagophore in S. cerevisiae

EMDB-15545: 
In situ cryo-electron tomogram of a bulk autophagy phagophore in S. cerevisiae #2

EMDB-15546: 
In situ cryo-electron tomogram of a bulk autophagy autophagosome with END cargo in S. cerevisiae #1

EMDB-15547: 
In situ cryo-electron tomogram of a bulk autophagy autophagosome fusing with the vacuole in S. cerevisiae #1

EMDB-15548: 
In situ cryo-electron tomogram of a bulk autophagy phagophore in S. cerevisiae #3

EMDB-15549: 
In situ cryo-electron tomogram of a bulk autophagy phagophore in S. cerevisiae #4

EMDB-31495: 
Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in LMNG/CHS detergents at pH ~8.0

EMDB-31496: 
Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in nanodiscs with soybean lipids at pH ~8.0

EMDB-31497: 
Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel with two conformationally different hemichannels

EMDB-14323: 
Structure of Chelator-GIDSR4 bound to Mdh2

EMDB-14324: 
Structure of Cage-GIDSR4 bound to PHSVTP-Fbp1

EMDB-14338: 
Structure of endogenous Cage-GIDAnt complex

EMDB-32830: 
GID subcomplex: Gid12 bound Substrate Receptor Scaffolding module

EMDB-32831: 
Gid12 bound GIDSR4 E3 ubiquitin ligase complex
EMDB-32833: 
Gid12 bound Chelator-GIDSR4

EMDB-32834: 
Cage assembly GID E3 ubiquitin ligase

EMDB-32835: 
Gid12 bound Cage-GIDSR3

EMDB-24822: 
20S proteasome from red blood cell lysate

EMDB-22995: 
Spike protein trimer

EMDB-23589: 
Cryo-EM structure of 2909 Fab in complex with 3BNC117 Fab and CAP256.wk34.c80 SOSIP.RnS2 N160K HIV-1 Env trimer

EMDB-23494: 
Cryo-EM of the SLFN12-PDE3A complex: PDE3A body refinement
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
