-Search query
-Search result
Showing 1 - 50 of 76 items for (author: lammens & k)

EMDB-19798: 
human PLD3 homodimer structure

PDB-8s86: 
human PLD3 homodimer structure

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template

PDB-8sxt: 
Structure of LINE-1 ORF2p with template:primer hybrid

PDB-8sxu: 
Structure of LINE-1 ORF2p with an oligo(A) template

EMDB-14762: 
Mot1:TBP:DNA - pre-hydrolysis state

PDB-7zke: 
Mot1:TBP:DNA - pre-hydrolysis state

EMDB-14584: 
Mot1(1-1836):TBP:DNA - post-hydrolysis complex dimer

PDB-7zb5: 
Mot1(1-1836):TBP:DNA - post-hydrolysis complex dimer
EMDB-14534: 
Mot1E1434Q:TBP:DNA - substrate recognition state

EMDB-14554: 
Mot1:TBP - product state

EMDB-14562: 
Mot1:TBP:DNA - post hydrolysis state

PDB-7z7n: 
Mot1E1434Q:TBP:DNA - substrate recognition state

PDB-7z8s: 
Mot1:TBP:DNA - post hydrolysis state

EMDB-14878: 
CtMre11-Rad50-Nbs1 complex, long coiled-coils map

EMDB-14879: 
Mre11-Rad50-Nbs1 complex (Chaetomium thermophilum) lower coiled-coils
EMDB-14880: 
Eukaryotic Mre11-Rad50-Nbs1 complex catalytic head

EMDB-14881: 
Chaetomium thermophilum Mre11-Rad50-Nbs1 complex bound to ATPyS (composite structure)

EMDB-15948: 
Human Mre11-Nbs1 complex

PDB-7zr1: 
Chaetomium thermophilum Mre11-Rad50-Nbs1 complex bound to ATPyS (composite structure)

PDB-8bah: 
Human Mre11-Nbs1 complex

EMDB-14877: 
Chaetomium thermophilum MRN complex (Nbs1)

EMDB-14882: 
Chaetomium thermophilum Rad50 zinc-hook tetramer

EMDB-14876: 
Cryo-EM map of Chaetomium thermophilum MRN (RBD & bridge)
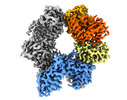
EMDB-14690: 
Human SLFN11 dimer apoenzyme

EMDB-14691: 
Human SLFN11 E209A dimer

EMDB-14692: 
Human SLFN11 dimer bound to ssDNA
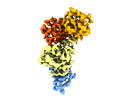
EMDB-14693: 
SLFN11 E209A monomer

EMDB-14695: 
SLFN11 dimer bound to tRNA

PDB-7zel: 
Human SLFN11 dimer apoenzyme

PDB-7zep: 
Human SLFN11 E209A dimer

PDB-7zes: 
Human SLFN11 dimer bound to ssDNA

EMDB-14391: 
Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA

EMDB-14392: 
Composite map of two E. coli Mre11-Rad50 (SbcCD) complexes bound to Ku70/80 blocked dsDNA in endonuclease state

EMDB-14393: 
Hairpin-bound state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and a DNA hairpin

EMDB-14394: 
C. thermophilum Ku70/80 heterodimer bound to DNA

EMDB-14403: 
Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and extended dsDNA

PDB-7yzo: 
Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA

PDB-7yzp: 
Hairpin-bound state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and a DNA hairpin

PDB-7z03: 
Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and extended dsDNA

EMDB-13581: 
human SLFN5

PDB-7ppj: 
human SLFN5

EMDB-12343: 
human ATM kinase with bound inhibitor KU-55933

EMDB-12345: 
human ATM Pincer domain

EMDB-12346: 
Human ATM Kinase Pincer-Spiral domains

EMDB-12347: 
Human ATM Spiral domain

EMDB-12350: 
Human ATM kinase domain with bound M4076 inhibitor

EMDB-12351: 
Human ATM kinase with bound inhibitor KU-55933 (composite structure)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
