+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human SLFN11 dimer bound to ssDNA | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | tRNA endonuclease / single-strand DNA binding protein / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationreplication fork arrest / negative regulation of G1/S transition of mitotic cell cycle / negative regulation of DNA replication / site of DNA damage / immune system process / helicase activity / endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / tRNA binding ...replication fork arrest / negative regulation of G1/S transition of mitotic cell cycle / negative regulation of DNA replication / site of DNA damage / immune system process / helicase activity / endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / tRNA binding / chromatin remodeling / DNA damage response / ATP hydrolysis activity / DNA binding / nucleoplasm / ATP binding / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
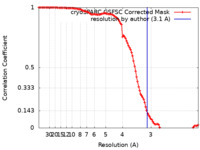
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Metzner FJ / Kugler M / Wenzl SJ / Lammens K | |||||||||
| Funding support |  Germany, European Union, 2 items Germany, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Mechanistic understanding of human SLFN11. Authors: Felix J Metzner / Simon J Wenzl / Michael Kugler / Stefan Krebs / Karl-Peter Hopfner / Katja Lammens /  Abstract: Schlafen 11 (SLFN11) is an interferon-inducible antiviral restriction factor with tRNA endoribonuclease and DNA binding functions. It is recruited to stalled replication forks in response to ...Schlafen 11 (SLFN11) is an interferon-inducible antiviral restriction factor with tRNA endoribonuclease and DNA binding functions. It is recruited to stalled replication forks in response to replication stress and inhibits replication of certain viruses such as the human immunodeficiency virus 1 (HIV-1) by modulating the tRNA pool. SLFN11 has been identified as a predictive biomarker in cancer, as its expression correlates with a beneficial response to DNA damage inducing anticancer drugs. However, the mechanism and interdependence of these two functions are largely unknown. Here, we present cryo-electron microscopy (cryo-EM) structures of human SLFN11 in its dimeric apoenzyme state, bound to tRNA and in complex with single-strand DNA. Full-length SLFN11 neither hydrolyses nor binds ATP and the helicase domain appears in an autoinhibited state. Together with biochemical and structure guided mutagenesis studies, our data give detailed insights into the mechanism of endoribonuclease activity as well as suggestions on how SLFN11 may block stressed replication forks. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14692.map.gz emd_14692.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14692-v30.xml emd-14692-v30.xml emd-14692.xml emd-14692.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14692_fsc.xml emd_14692_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_14692.png emd_14692.png | 115.5 KB | ||
| Masks |  emd_14692_msk_1.map emd_14692_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14692.cif.gz emd-14692.cif.gz | 6.1 KB | ||
| Others |  emd_14692_half_map_1.map.gz emd_14692_half_map_1.map.gz emd_14692_half_map_2.map.gz emd_14692_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14692 http://ftp.pdbj.org/pub/emdb/structures/EMD-14692 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14692 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14692 | HTTPS FTP |
-Related structure data
| Related structure data |  7zesMC  7zelC  7zepC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14692.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14692.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.046 Å | ||||||||||||||||||||||||||||||||||||
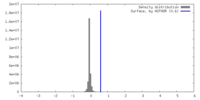
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14692_msk_1.map emd_14692_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
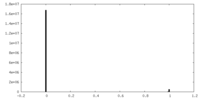
| Density Histograms |
-Half map: half map A
| File | emd_14692_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
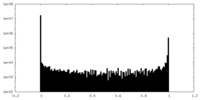
| Density Histograms |
-Half map: half map B
| File | emd_14692_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
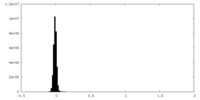
| Density Histograms |
- Sample components
Sample components
-Entire : SLFN11 dimer bound to ssDNA
| Entire | Name: SLFN11 dimer bound to ssDNA |
|---|---|
| Components |
|
-Supramolecule #1: SLFN11 dimer bound to ssDNA
| Supramolecule | Name: SLFN11 dimer bound to ssDNA / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: Schlafen family member 11
| Supramolecule | Name: Schlafen family member 11 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: DNA (5'-D(P*CP*GP*CP*GP*T)-3')
| Supramolecule | Name: DNA (5'-D(P*CP*GP*CP*GP*T)-3') / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) / Synthetically produced: Yes |
-Macromolecule #1: Schlafen family member 11
| Macromolecule | Name: Schlafen family member 11 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: Hydrolases; Acting on acid anhydrides |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 106.207914 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MADYKDDDDK GTDYKDDDDK LEVLFQGPME ANQCPLVVEP SYPDLVINVG EVTLGEENRK KLQKIQRDQE KERVMRAACA LLNSGGGVI RMAKKVEHPV EMGLDLEQSL RELIQSSDLQ AFFETKQQGR CFYIFVKSWS SGPFPEDRSV KPRLCSLSSS L YRRSETSV ...String: MADYKDDDDK GTDYKDDDDK LEVLFQGPME ANQCPLVVEP SYPDLVINVG EVTLGEENRK KLQKIQRDQE KERVMRAACA LLNSGGGVI RMAKKVEHPV EMGLDLEQSL RELIQSSDLQ AFFETKQQGR CFYIFVKSWS SGPFPEDRSV KPRLCSLSSS L YRRSETSV RSMDSREAFC FLKTKRKPKI LEEGPFHKIH KGVYQELPNS DPADPNSDPA DLIFQKDYLE YGEILPFPES QL VEFKQFS TKHFQEYVKR TIPEYVPAFA NTGGGYLFIG VDDKSREVLG CAKENVDPDS LRRKIEQAIY KLPCVHFCQP QRP ITFTLK IVNVLKRGEL YGYACMIRVN PFCCAVFSEA PNSWIVEDKY VCSLTTEKWV GMMTDTDPDL LQLSEDFECQ LSLS SGPPL SRPVYSKKGL EHKKELQQLL FSVPPGYLRY TPESLWRDLI SEHRGLEELI NKQMQPFFRG ILIFSRSWAV DLNLQ EKPG VICDALLIAQ NSTPILYTIL REQDAEGQDY CTRTAFTLKQ KLVNMGGYTG KVCVRAKVLC LSPESSAEAL EAAVSP MDY PASYSLAGTQ HMEALLQSLV IVLLGFRSLL SDQLGCEVLN LLTAQQYEIF SRSLRKNREL FVHGLPGSGK TIMAMKI ME KIRNVFHCEA HRILYVCENQ PLRNFISDRN ICRAETRKTF LRENFEHIQH IVIDEAQNFR TEDGDWYGKA KSITRRAK G GPGILWIFLD YFQTSHLDCS GLPPLSDQYP REELTRIVRN ADPIAKYLQK EMQVIRSNPS FNIPTGCLEV FPEAEWSQG VQGTLRIKKY LTVEQIMTCV ADTCRRFFDR GYSPKDVAVL VSTAKEVEHY KYELLKAMRK KRVVQLSDAC DMLGDHIVLD SVRRFSGLE RSIVFGIHPR TADPAILPNV LICLASRAKQ HLYIFPWGGH UniProtKB: Schlafen family member 11 |
-Macromolecule #2: DNA (5'-D(P*CP*GP*CP*GP*T)-3')
| Macromolecule | Name: DNA (5'-D(P*CP*GP*CP*GP*T)-3') / type: dna / ID: 2 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 1.496011 KDa |
| Sequence | String: (DC)(DG)(DC)(DG)(DT) |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 43.33 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.9 µm / Nominal defocus min: 1.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)