+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SLFN11 E209A monomer | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
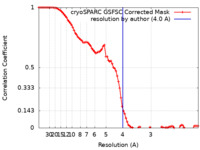
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Metzner FJ / Kugler M / Wenzl SJ / Lammens K | |||||||||
| Funding support |  Germany, European Union, 2 items Germany, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Mechanistic understanding of human SLFN11. Authors: Felix J Metzner / Simon J Wenzl / Michael Kugler / Stefan Krebs / Karl-Peter Hopfner / Katja Lammens /  Abstract: Schlafen 11 (SLFN11) is an interferon-inducible antiviral restriction factor with tRNA endoribonuclease and DNA binding functions. It is recruited to stalled replication forks in response to ...Schlafen 11 (SLFN11) is an interferon-inducible antiviral restriction factor with tRNA endoribonuclease and DNA binding functions. It is recruited to stalled replication forks in response to replication stress and inhibits replication of certain viruses such as the human immunodeficiency virus 1 (HIV-1) by modulating the tRNA pool. SLFN11 has been identified as a predictive biomarker in cancer, as its expression correlates with a beneficial response to DNA damage inducing anticancer drugs. However, the mechanism and interdependence of these two functions are largely unknown. Here, we present cryo-electron microscopy (cryo-EM) structures of human SLFN11 in its dimeric apoenzyme state, bound to tRNA and in complex with single-strand DNA. Full-length SLFN11 neither hydrolyses nor binds ATP and the helicase domain appears in an autoinhibited state. Together with biochemical and structure guided mutagenesis studies, our data give detailed insights into the mechanism of endoribonuclease activity as well as suggestions on how SLFN11 may block stressed replication forks. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14693.map.gz emd_14693.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14693-v30.xml emd-14693-v30.xml emd-14693.xml emd-14693.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14693_fsc.xml emd_14693_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_14693.png emd_14693.png | 55.2 KB | ||
| Masks |  emd_14693_msk_1.map emd_14693_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_14693_half_map_1.map.gz emd_14693_half_map_1.map.gz emd_14693_half_map_2.map.gz emd_14693_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14693 http://ftp.pdbj.org/pub/emdb/structures/EMD-14693 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14693 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14693 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14693.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14693.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.046 Å | ||||||||||||||||||||||||||||||||||||
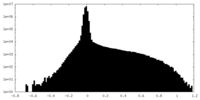
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14693_msk_1.map emd_14693_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_14693_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_14693_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SLFN11 E209A monomer
| Entire | Name: SLFN11 E209A monomer |
|---|---|
| Components |
|
-Supramolecule #1: SLFN11 E209A monomer
| Supramolecule | Name: SLFN11 E209A monomer / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Macromolecule #1: SLFN11 E209A
| Macromolecule | Name: SLFN11 E209A / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MADYKDDDDK GTDYKDDDDK LEVLFQGPME ANQCPLVVEP SYPDLVINVG EVTLGEENRK KLQKIQRDQE KERVMRAACA LLNSGGGVIR MAKKVEHPVE MGLDLEQSLR ELIQSSDLQA FFETKQQGRC FYIFVKSWSS GPFPEDRSVK PRLCSLSSSL YRRSETSVRS ...String: MADYKDDDDK GTDYKDDDDK LEVLFQGPME ANQCPLVVEP SYPDLVINVG EVTLGEENRK KLQKIQRDQE KERVMRAACA LLNSGGGVIR MAKKVEHPVE MGLDLEQSLR ELIQSSDLQA FFETKQQGRC FYIFVKSWSS GPFPEDRSVK PRLCSLSSSL YRRSETSVRS MDSREAFCFL KTKRKPKILE EGPFHKIHKG VYQELPNSDP ADPNSDPADL IFQKDYLEYG EILPFPASQL VEFKQFSTKH FQEYVKRTIP EYVPAFANTG GGYLFIGVDD KSREVLGCAK ENVDPDSLRR KIEQAIYKLP CVHFCQPQRP ITFTLKIVNV LKRGELYGYA CMIRVNPFCC AVFSEAPNSW IVEDKYVCSL TTEKWVGMMT DTDPDLLQLS EDFECQLSLS SGPPLSRPVY SKKGLEHKKE LQQLLFSVPP GYLRYTPESL WRDLISEHRG LEELINKQMQ PFFRGILIFS RSWAVDLNLQ EKPGVICDAL LIAQNSTPIL YTILREQDAE GQDYCTRTAF TLKQKLVNMG GYTGKVCVRA KVLCLSPESS AEALEAAVSP MDYPASYSLA GTQHMEALLQ SLVIVLLGFR SLLSDQLGCE VLNLLTAQQY EIFSRSLRKN RELFVHGLPG SGKTIMAMKI MEKIRNVFHC EAHRILYVCE NQPLRNFISD RNICRAETRK TFLRENFEHI QHIVIDEAQN FRTEDGDWYG KAKSITRRAK GGPGILWIFL DYFQTSHLDC SGLPPLSDQY PREELTRIVR NADPIAKYLQ KEMQVIRSNP SFNIPTGCLE VFPEAEWSQG VQGTLRIKKY LTVEQIMTCV ADTCRRFFDR GYSPKDVAVL VSTAKEVEHY KYELLKAMRK KRVVQLSDAC DMLGDHIVLD SVRRFSGLER SIVFGIHPRT ADPAILPNVL ICLASRAKQH LYIFPWGGH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 43.58 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.9 µm / Nominal defocus min: 1.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)