+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mot1:TBP - product state | |||||||||
 Map data Map data | main map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Woike S / Eustermann S / Jung J / Wenzl SJ / Hagemann G / Bartho JD / Lammens K / Butryn A / Herzog F / Hopfner K-P | |||||||||
| Funding support |  Germany, European Union, 2 items Germany, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1. Authors: Stephan Woike / Sebastian Eustermann / James Jung / Simon Josef Wenzl / Götz Hagemann / Joseph Bartho / Katja Lammens / Agata Butryn / Franz Herzog / Karl-Peter Hopfner /     Abstract: The Swi2/Snf2 family transcription regulator Modifier of Transcription 1 (Mot1) uses adenosine triphosphate (ATP) to dissociate and reallocate the TATA box-binding protein (TBP) from and between ...The Swi2/Snf2 family transcription regulator Modifier of Transcription 1 (Mot1) uses adenosine triphosphate (ATP) to dissociate and reallocate the TATA box-binding protein (TBP) from and between promoters. To reveal how Mot1 removes TBP from TATA box DNA, we determined cryogenic electron microscopy structures that capture different states of the remodeling reaction. The resulting molecular video reveals how Mot1 dissociates TBP in a process that, intriguingly, does not require DNA groove tracking. Instead, the motor grips DNA in the presence of ATP and swings back after ATP hydrolysis, moving TBP to a thermodynamically less stable position on DNA. Dislodged TBP is trapped by a chaperone element that blocks TBP's DNA binding site. Our results show how Swi2/Snf2 proteins can remodel protein-DNA complexes through DNA bending without processive DNA tracking and reveal mechanistic similarities to RNA gripping DEAD box helicases and RIG-I-like immune sensors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14554.map.gz emd_14554.map.gz | 79 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14554-v30.xml emd-14554-v30.xml emd-14554.xml emd-14554.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14554_fsc.xml emd_14554_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_14554.png emd_14554.png | 47.3 KB | ||
| Masks |  emd_14554_msk_1.map emd_14554_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Others |  emd_14554_half_map_1.map.gz emd_14554_half_map_1.map.gz emd_14554_half_map_2.map.gz emd_14554_half_map_2.map.gz | 77.7 MB 77.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14554 http://ftp.pdbj.org/pub/emdb/structures/EMD-14554 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14554 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14554 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14554.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14554.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||
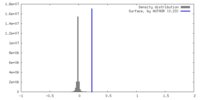
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14554_msk_1.map emd_14554_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
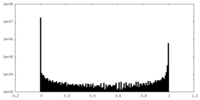
| Density Histograms |
-Half map: half map A
| File | emd_14554_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
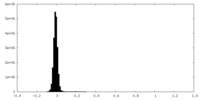
| Density Histograms |
-Half map: half map B
| File | emd_14554_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
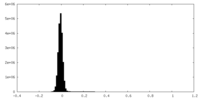
| Density Histograms |
- Sample components
Sample components
-Entire : Mot1:TBP complex after DNA release
| Entire | Name: Mot1:TBP complex after DNA release |
|---|---|
| Components |
|
-Supramolecule #1: Mot1:TBP complex after DNA release
| Supramolecule | Name: Mot1:TBP complex after DNA release / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
-Macromolecule #1: Mot1:TBP complex
| Macromolecule | Name: Mot1:TBP complex / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
| Recombinant expression | Organism:  |
| Sequence | String: MATRLDRLVT ILETGSTRLI RDTAVNQLAD WQKQHPEELF NLLSRVVPYL RHKDWETRTT AAKAIGKIIE NAPLYDPNAG QDEAAPEPTN GSFEVKKEEE KDVLEQDNFF RLESLDVATI VKYGRPLLRG GPVDYNLAAL DPQKRLAHLK KTLNGRLGLL GRVFEDEEMP ...String: MATRLDRLVT ILETGSTRLI RDTAVNQLAD WQKQHPEELF NLLSRVVPYL RHKDWETRTT AAKAIGKIIE NAPLYDPNAG QDEAAPEPTN GSFEVKKEEE KDVLEQDNFF RLESLDVATI VKYGRPLLRG GPVDYNLAAL DPQKRLAHLK KTLNGRLGLL GRVFEDEEMP VEQIASPITP NDAAGANGVG RQDGASNDNQ SQAIDESKMS ARQLNVLKRK RKREAQKAAQ GKSGFGDLSL RRSTTAGSDA FGEDTPMPDA DSKKNKLAEY FSLDRPENTE EDTKIVSEFK GPVLPIKSEI EADDSLEGAE WPFERLCEFL KVDLFDPQWE TRHGAAMGLR EVIRVHGAGA GRRRGKTRKE NNDLNRQWLD DLAYRLLCVL MLDKFTDYSS DTSVAPIRET VGQTLGAVLR HISVESVHAI YRLLYCMVTQ EDLPSEQNMW AVCHGGMVGL RYVVAVRKDL LLQDGDMIDG VVRCVMQGLG DIDDDVRSVS AATLIPMAKE FVMMRRSALD SLINIVWESL SNLGDDLSAS TGKIMDLLAT LCSFPEVLEA MKVSASQDEE RSFTLLVPRL YPFLRHTITS VRLAVLKALM TFANLGGETS QGWLNGRILR LIFQNIIVER DQDTLNMSLE LWTTLVRRLA ARDPAILADE FEAHAEPMMQ LALHPIGVPR HPIPMNPALF QKPSGGTYSL PGASQTNSRR SSPPEGERAT KRRRKSTKAE DVAPSTHTHD VDGHMIQGEV DLVGVDVLIR SRISAAKAMG LIMSFIPTPR LASYDTAVLQ ALSSPYASTQ LAAAMVIDEY AKNCSTPEVA SRFIEPLQKI IDLERPSHYR DLVTYVQRVR SASQQLINLF RDHGKVSQGK LPTLAVVVQG EPEAGPGAFS IANAEKVVNE DFERLKRLMA PGQRLIALPQ LNEAREQTVE VIEEAKAAKE ARDARIKAAA ACALVAMKVL PKKPSPLIKA IMDSIKTEEN QELQSRSAAT IARLVQLFTE SGRRGPAEKV VANLVKFSCV EVAETPEFPI HAHKTNVILS MQKEEDRVDH PDAVKYAREA KAARITRRGA KEALEILSKN FGAELLERVP TLRTFMEEPL VRAFSGDLPP EARDPENAFG QEIVDAMSVI RTMTPTLHPA LHPFVMQQVP LVIKALRSDL SVFRYMAAKC MATICSVITV DGMTALVEKV LPSINNPLDL SFRQGAIEVI YHLIAVMGDA ILPYVIFLIV PVLGRMSDSD NQIRLIATTS FATLVKLVPL EAGIPDPPGL SEELLKGRDR ERTFIAQLLD PKKIEPFKIP VAIKAELRSY QQEGVNWLAF LNKYHLHGIL CDDMGLGKTL QTICIVASDH HQRAEEFART GAPEVRKLPS LIICPPTLSG HWQQEIKTYA PFLTVTAYVG SPAERRAMKD SLDKTDIVIT SYDVCRNDID VIEKYNWNYC VLDEGHLIKN PKAKITLAVK RLTSNHRLIL TGTPIQNNVL ELWSLFDFLM PGFLGAEKVF LDRFAKPIAN SRYSKASSKE QEAGALAIEA LHKQVLPFLL RRLKEEVLND LPPKILQNYY CDLSDLQRKL FEDFTKREGK KITETAGRDD KEAKQHIFQA LQYMRKLCNS PALVMKPGHK AYEDTQKYLA KHGTTLEDPI HAPKLGALRD LLVDCGIGVE GQESSDPLYT PIKPHRALIF CQMKEMLDMV QNTVLKQMLP SVSYLRLDGS VEANKRQDIV NKFNSDPSYD VLLLTTSVGG LGLNLTGADT VIFVEHDWNP QKDLQAMDRA HRIGQKKVVN VYRIITRGTL EEKILSLQRF KIDVASTVVN QQNAGLATMD TDQILDLFNL GESGPSLITD NKESIEGREE DMVDIETGDV RRPGKKAAWL EGLGELWDNA QYEESFDLDG FLKTMQAAAW SHPQFEK MG SSHHHHHHSS GENLYFQGHM EAIQTHPANA AQAKAFTAPG SLSFPGGASE IANSTAPT N GASNGGQQQG VQATSGAGVT PATPAATPGA GPAGPSGITP TLQNIVATVN LDCRLDLKT IALHARNAEY NPKRFAAVIM RIREPKTTAL IFASGKMVVT GAKSEDDSKL ASRKYARIIQ KLGFNAKFT DFKIQNIVGS CDIKFPIRLE GLASKHHNFS SYEPELFPGL IYRMIKPKIV L LIFVSGKI VLTGAKVREE IYQAFEMIYP VLQDFRKV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Incubation with 1 mM ATP-gamma-S before SEC and before grid preparation. 0.05% beta-octyl glucoside added before blotting. | ||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 7 sec. / Pretreatment - Atmosphere: AIR | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 283 K / Instrument: LEICA EM GP Details: Incubation on the grid for 20 seconds before plunging.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)