-Search query
-Search result
Showing 1 - 50 of 66 items for (author: lambe & t)

EMDB-42371: 
Mouse apoferritin imaged with a square aperture

EMDB-42372: 
Mouse apoferritin imaged with a round aperture

EMDB-42373: 
Mouse apoferritin imaged with a square aperture with P2 projection lens rotation, reconstructed from 25,000 particles

EMDB-42374: 
Mouse apoferritin imaged with a round aperture without P2 projection lens rotation, reconstructed from 25,000 particles

EMDB-16820: 
Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 1).

EMDB-16821: 
Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 2).

EMDB-16822: 
Cryo-EM structure of the murine IL-12 complete extracellular signaling complex (Class 1).

EMDB-16823: 
Cryo-EM structure of the murine IL-12 complete extracellular signaling complex (Class 2).

EMDB-16824: 
Cryo-EM structure of a pre-dimerized human IL-23 complete extracellular signaling complex.

EMDB-17580: 
Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 1), obtained after local refinement.

PDB-8odz: 
Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 1).

PDB-8oe0: 
Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 2).

PDB-8oe4: 
Cryo-EM structure of a pre-dimerized human IL-23 complete extracellular signaling complex.

PDB-8pb1: 
Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 1), obtained after local refinement.
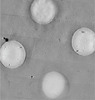
EMDB-42851: 
5x5 tiled montage tomogram of a holey carbon grid with apoferritin imaged with a square electron beam

EMDB-42879: 
3x3 tiled montage tomogram of a yeast lamella imaged with a square electron beam
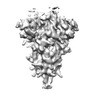
EMDB-16403: 
Subtomogram averaging of a coronavirus spike (ChAdOx1 19E6) in C3 symmetry
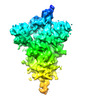
EMDB-16404: 
Subtomogram averaging of a coronavirus spike (ChAdOx1 19E6) in C1 symmetry

EMDB-16405: 
Subtomogram averaging of a coronavirus spike (ChAdOx1 19E) in C3 symmetry

EMDB-16406: 
Subtomogram averaging of a coronavirus spike in situ (ChAdOx1 19E) in C1 symmetry

EMDB-16697: 
Subtomogram averaging of a coronavirus spike (ChAdOx1 19E6) in C1 symmetry after 3D classification

EMDB-14717: 
SARS CoV Spike protein, Closed C3 conformation

EMDB-14718: 
SARS CoV Spike protein, Closed C1 conformation

EMDB-14724: 
SARS CoV Spike protein, Open conformation

PDB-7zh1: 
SARS CoV Spike protein, Closed C3 conformation

PDB-7zh2: 
SARS CoV Spike protein, Closed C1 conformation

PDB-7zh5: 
SARS CoV Spike protein, Open conformation

EMDB-13608: 
Focused reconstruction of Haliangium ochraceum encapsulated ferritin cargo within the encapsulin nano compartment
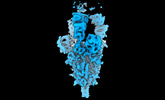
EMDB-14885: 
OMI-42 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN

EMDB-14886: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE RBD (local refinement)

EMDB-14887: 
OMI-2 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN
EMDB-14910: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE

PDB-7zr7: 
OMI-42 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN

PDB-7zr8: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE RBD (local refinement)

PDB-7zr9: 
OMI-2 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN

PDB-7zrc: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE

EMDB-12853: 
Icosahedral cryo-EM reconstruction of Haliangium ochraceum encapsulin

PDB-7odw: 
Model of Haliangium ochraceum encapsulin from icosahedral single particle reconstruction

EMDB-12859: 
Model of closed pentamer of the Haliangium ochraceum encapsulin from symmetry expansion of icosahedral single particle reconstruction

EMDB-12864: 
Single particle reconstruction of open pentamer of the Haliangium ochraceum encapsulin from symmetry expansion of icosahedral single particle reconstruction

EMDB-12873: 
Asymmetric single particle reconstruction of the Haliangium ochraceum encapsulin encapsulated ferritin complex

EMDB-13603: 
Asymmetric single particle reconstruction of the Haliangium ochraceum encapsulin encapsulated ferritin complex calculated using Cryosparc

PDB-7oe2: 
Model of closed pentamer of the Haliangium ochraceum encapsulin from symmetry expansion of icosahedral single particle reconstruction

PDB-7oeu: 
Model of open pentamer of the Haliangium ochraceum encapsulin from symmetry expansion of icosahedral single particle reconstruction

EMDB-12530: 
Subtomogram average of ChAdOx1 nCoV-19/AZD1222 derived SARS-CoV-2 spike glycoprotein

EMDB-10371: 
CryoEM structure of MexB in nanodisc

EMDB-10372: 
CryoEM structure of MexAB-OprM in nanodisc

EMDB-10395: 
MexA-MexB cryoEM map

PDB-6t7s: 
MexB structure solved by cryo-EM in nanodisc in absence of its protein partners

PDB-6ta5: 
OprM-MexA complex from the MexAB-OprM Pseudomonas aeruginosa whole assembly reconstituted in nanodiscs
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
