[English] 日本語
 Yorodumi
Yorodumi- EMDB-16822: Cryo-EM structure of the murine IL-12 complete extracellular sign... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the murine IL-12 complete extracellular signaling complex (Class 1). | |||||||||
 Map data Map data | Sharpened map of the murine IL12:IL12Rbeta1:IL12Rbeta2 complex (Class 1). | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Cytokine / Receptor / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationinterleukin-12 beta subunit binding / Interleukin-12 signaling / Interleukin-23 signaling / interleukin-23 receptor binding / Interleukin-35 Signalling / interleukin-12 alpha subunit binding / interleukin-12 complex / interleukin-23 complex / T-helper 1 cell activation / negative regulation of vascular endothelial growth factor signaling pathway ...interleukin-12 beta subunit binding / Interleukin-12 signaling / Interleukin-23 signaling / interleukin-23 receptor binding / Interleukin-35 Signalling / interleukin-12 alpha subunit binding / interleukin-12 complex / interleukin-23 complex / T-helper 1 cell activation / negative regulation of vascular endothelial growth factor signaling pathway / positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target / negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis / positive regulation of T-helper 1 type immune response / positive regulation of smooth muscle cell apoptotic process / interleukin-12 receptor binding / interleukin-23 receptor complex / T-helper cell differentiation / interleukin-12-mediated signaling pathway / positive regulation of NK T cell proliferation / negative regulation of interleukin-17 production / positive regulation of osteoclast differentiation / cytokine receptor activity / natural killer cell activation / positive regulation of granulocyte macrophage colony-stimulating factor production / response to UV-B / positive regulation of T cell differentiation / negative regulation of interleukin-10 production / defense response to protozoan / positive regulation of interleukin-17 production / positive regulation of natural killer cell proliferation / positive regulation of activated T cell proliferation / positive regulation of interleukin-10 production / negative regulation of protein secretion / cell surface receptor signaling pathway via JAK-STAT / T cell proliferation / coreceptor activity / positive regulation of defense response to virus by host / response to cytokine / positive regulation of interleukin-12 production / positive regulation of T cell proliferation / cytokine activity / endosome lumen / growth factor activity / negative regulation of inflammatory response to antigenic stimulus / negative regulation of smooth muscle cell proliferation / positive regulation of T cell mediated cytotoxicity / Golgi lumen / cellular response to type II interferon / positive regulation of type II interferon production / positive regulation of tumor necrosis factor production / cell migration / cellular response to lipopolysaccharide / response to lipopolysaccharide / defense response to virus / defense response to Gram-negative bacterium / cell surface receptor signaling pathway / cell population proliferation / immune response / endoplasmic reticulum lumen / protein heterodimerization activity / external side of plasma membrane / protein kinase binding / cell surface / extracellular space / extracellular region / identical protein binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.6 Å | |||||||||
 Authors Authors | Felix J / Bloch Y / Savvides SN | |||||||||
| Funding support |  Belgium, 2 items Belgium, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23. Authors: Yehudi Bloch / Jan Felix / Romain Merceron / Mathias Provost / Royan Alipour Symakani / Robin De Backer / Elisabeth Lambert / Ahmad R Mehdipour / Savvas N Savvides /     Abstract: Cell-surface receptor complexes mediated by pro-inflammatory interleukin (IL)-12 and IL-23, both validated therapeutic targets, are incompletely understood due to the lack of structural insights into ...Cell-surface receptor complexes mediated by pro-inflammatory interleukin (IL)-12 and IL-23, both validated therapeutic targets, are incompletely understood due to the lack of structural insights into their complete extracellular assemblies. Furthermore, there is a paucity of structural details describing the IL-12-receptor interaction interfaces, in contrast to IL-23-receptor complexes. Here we report structures of fully assembled mouse IL-12/human IL-23-receptor complexes comprising the complete extracellular segments of the cognate receptors determined by electron cryo-microscopy. The structures reveal key commonalities but also surprisingly diverse features. Most notably, whereas IL-12 and IL-23 both utilize a conspicuously presented aromatic residue on their α-subunit as a hotspot to interact with the N-terminal Ig domain of their high-affinity receptors, only IL-12 juxtaposes receptor domains proximal to the cell membrane. Collectively, our findings will help to complete our understanding of cytokine-mediated assemblies of tall cytokine receptors and will enable a cytokine-specific interrogation of IL-12/IL-23 signaling in physiology and disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16822.map.gz emd_16822.map.gz | 38.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16822-v30.xml emd-16822-v30.xml emd-16822.xml emd-16822.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16822_fsc.xml emd_16822_fsc.xml | 7.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_16822.png emd_16822.png | 78.8 KB | ||
| Masks |  emd_16822_msk_1.map emd_16822_msk_1.map | 40.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16822.cif.gz emd-16822.cif.gz | 6.6 KB | ||
| Others |  emd_16822_half_map_1.map.gz emd_16822_half_map_1.map.gz emd_16822_half_map_2.map.gz emd_16822_half_map_2.map.gz | 37.7 MB 37.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16822 http://ftp.pdbj.org/pub/emdb/structures/EMD-16822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16822 | HTTPS FTP |
-Related structure data
| Related structure data |  8c7mC  8cr5C  8cr6C  8cr8C  8odxC  8odzC  8oe0C  8oe4C  8pb1C  8ppmC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16822.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16822.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of the murine IL12:IL12Rbeta1:IL12Rbeta2 complex (Class 1). | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.51 Å | ||||||||||||||||||||||||||||||||||||
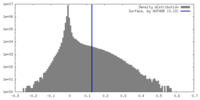
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16822_msk_1.map emd_16822_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
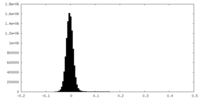
| Density Histograms |
-Half map: Half map 2 of the murine IL12:IL12Rbeta1:IL12Rbeta2 complex...
| File | emd_16822_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of the murine IL12:IL12Rbeta1:IL12Rbeta2 complex (Class 1). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 of the murine IL12:IL12Rbeta1:IL12Rbeta2 complex...
| File | emd_16822_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of the murine IL12:IL12Rbeta1:IL12Rbeta2 complex (Class 1). | ||||||||||||
| Projections & Slices |
| ||||||||||||
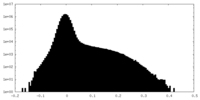
| Density Histograms |
- Sample components
Sample components
-Entire : Murine IL-12 in complex with Strep-II tagged mIL-12Rbeta1 and mIL...
| Entire | Name: Murine IL-12 in complex with Strep-II tagged mIL-12Rbeta1 and mIL-12Rbeta2. |
|---|---|
| Components |
|
-Supramolecule #1: Murine IL-12 in complex with Strep-II tagged mIL-12Rbeta1 and mIL...
| Supramolecule | Name: Murine IL-12 in complex with Strep-II tagged mIL-12Rbeta1 and mIL-12Rbeta2. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 198 KDa |
-Macromolecule #1: Murine Interleukin-12 subunit alpha (IL-12A, p35), with a C-termi...
| Macromolecule | Name: Murine Interleukin-12 subunit alpha (IL-12A, p35), with a C-terminal caspase-3 cleavable His tag. type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MCQSRYLLFL ATLALLNHLS LARVIPVSGP ARCLSQSRNL LKTTDDMVKT AREKLKHYSC TAEDIDHEDI TRDQTSTLKT CLPLELHKNE SCLATRETSS TTRGSCLPPQ KTSLMMTLCL GSIYEDLKMY QTEFQAINAA LQNHNHQQII LDKGMLVAID ELMQSLNHNG ...String: MCQSRYLLFL ATLALLNHLS LARVIPVSGP ARCLSQSRNL LKTTDDMVKT AREKLKHYSC TAEDIDHEDI TRDQTSTLKT CLPLELHKNE SCLATRETSS TTRGSCLPPQ KTSLMMTLCL GSIYEDLKMY QTEFQAINAA LQNHNHQQII LDKGMLVAID ELMQSLNHNG ETLRQKPPVG EADPYRVKMK LCILLHAFST RVVTINRVMG YLSSAGTSDE VDGGSGGSGL NDIFEAQKIE WHEGRTKHHH HHH UniProtKB: Interleukin-12 subunit alpha |
-Macromolecule #2: Murine Interleukin-12 subunit beta (IL-12B, p40).
| Macromolecule | Name: Murine Interleukin-12 subunit beta (IL-12B, p40). / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MCPQKLTISW FAIVLLVSPL MAMWELEKDV YVVEVDWTPD APGETVNLTC DTPEEDDITW TSDQRHGVIG SGKTLTITVK EFLDAGQYTC HKGGETLSHS HLLLHKKENG IWSTEILKNF KNKTFLKCEA PNYSGRFTCS WLVQRNMDLK FNIKSSSSSP DSRAVTCGMA ...String: MCPQKLTISW FAIVLLVSPL MAMWELEKDV YVVEVDWTPD APGETVNLTC DTPEEDDITW TSDQRHGVIG SGKTLTITVK EFLDAGQYTC HKGGETLSHS HLLLHKKENG IWSTEILKNF KNKTFLKCEA PNYSGRFTCS WLVQRNMDLK FNIKSSSSSP DSRAVTCGMA SLSAEKVTLD QRDYEKYSVS CQEDVTCPTA EETLPIELAL EARQQNKYEN YSTSFFIRDI IKPDPPKNLQ MKPLKNSQVE VSWEYPDSWS TPHSYFSLKF FVRIQRKKEK MKETEEGCNQ KGAFLVEKTS TEVQCKGGNV CVQAQDRYYN SSCSKWACVP CRVRS UniProtKB: Interleukin-12 subunit beta |
-Macromolecule #3: Murine Interleukin-12 Receptor beta 1 (domains 1 to 5) with C-ter...
| Macromolecule | Name: Murine Interleukin-12 Receptor beta 1 (domains 1 to 5) with C-terminal Strep-II tag. type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QLGASGPGDG CCVEKTSFPE GASGSPLGPR NLSCYRVSKT DYECSWQYDG PEDNVSHVLW CCFVPPNHTH TGQERCRYFS SGPDRTVQFW EQDGIPVLSK VNFWVESRLG NRTMKSQKIS QYLYNWTKTT PPLGHIKVSQ SHRQLRMDWN VSEEAGAEVQ FRRRMPTTNW ...String: QLGASGPGDG CCVEKTSFPE GASGSPLGPR NLSCYRVSKT DYECSWQYDG PEDNVSHVLW CCFVPPNHTH TGQERCRYFS SGPDRTVQFW EQDGIPVLSK VNFWVESRLG NRTMKSQKIS QYLYNWTKTT PPLGHIKVSQ SHRQLRMDWN VSEEAGAEVQ FRRRMPTTNW TLGDCGPQVN SGSGVLGDIR GSMSESCLCP SENMAQEIQI RRRRRLSSGA PGGPWSDWSM PVCVPPEVLP QAKIKFLVEP LNQGGRRRLT MQGQSPQLAV PEGCRGRPGA QVKKHLVLVR MLSCRCQAQT SKTVPLGKKL NLSGATYDLN VLAKTRFGRS TIQKWHLPAQ ELTETRALNV SVGGNMTSMQ WAAQAPGTTY CLEWQPWFQH RNHTHCTLIV PEEEDPAKMV THSWSSKPTL EQEECYRITV FASKNPKNPM LWATVLSSYY FGGNASRAGT PRHVSVRNQT GDSVSVEWTA SQLSTCPGVL TQYVVRCEAE DGAWESEWLV PPTKTQVTLD GLRSRVMYKV QVRADTARLP GAWSHPQRFS FEGTWSHPQF EK UniProtKB: Interleukin-12 receptor subunit beta-1 |
-Macromolecule #4: Murine Interleukin-12 Receptor beta 2 (domains 1 to 6) with C-ter...
| Macromolecule | Name: Murine Interleukin-12 Receptor beta 2 (domains 1 to 6) with C-terminal Strep-II tag. type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NIDVCKLGTV TVQPAPVIPL GSAANISCSL NPKQGCSHYP SSNELILLKF VNDVLVENLH GKKVHDHTGH SSTFQVTNLS LGMTLFVCKL NCSNSQKKPP VPVCGVEISV GVAPEPPQNI SCVQEGENGT VACSWNSGKV TYLKTNYTLQ LSGPNNLTCQ KQCFSDNRQN ...String: NIDVCKLGTV TVQPAPVIPL GSAANISCSL NPKQGCSHYP SSNELILLKF VNDVLVENLH GKKVHDHTGH SSTFQVTNLS LGMTLFVCKL NCSNSQKKPP VPVCGVEISV GVAPEPPQNI SCVQEGENGT VACSWNSGKV TYLKTNYTLQ LSGPNNLTCQ KQCFSDNRQN CNRLDLGINL SPDLAESRFI VRVTAINDLG NSSSLPHTFT FLDIVIPLPP WDIRINFLNA SGSRGTLQWE DEGQVVLNQL RYQPLNSTSW NMVNATNAKG KYDLRDLRPF TEYEFQISSK LHLSGGSWSN WSESLRTRTP EEEPVGILDI WYMKQDIDYD RQQISLFWKS LNPSEARGKI LHYQVTLQEV TKKTTLQNTT RHTSWTRVIP RTGAWTASVS AANSKGASAP THINIVDLCG TGLLAPHQVS AKSENMDNIL VTWQPPKKAD SAVREYIVEW RALQPGSITK FPPHWLRIPP DNMSALISEN IKPYICYEIR VHALSESQGG CSSIRGDSKH KAPVSGPHIT AITEKKERLF ISWTHIPFPE QRGCILHYRI YWKERDSTAQ PELCEIQYRR SQNSHPISSL QPRVTYVLWM TAVTAAGESP QGNEREFCPQ GKANGTWSHP QFEK UniProtKB: Interleukin-12 receptor subunit beta-2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
Details: HEPES-buffered saline (HBS): 25 mM HEPES, pH 7.4, 150 mM NaCl. | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: LEICA PLUNGER / Details: Leica EM GP2, 4 s. blotting time.. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 7174 / Average exposure time: 3.37 sec. / Average electron dose: 61.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 60000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)