[English] 日本語
 Yorodumi
Yorodumi- EMDB-16824: Cryo-EM structure of a pre-dimerized human IL-23 complete extrace... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a pre-dimerized human IL-23 complete extracellular signaling complex. | |||||||||
 Map data Map data | Main map: Unfiltered, non-sharpened map of the human IL23:IL12Rbeta1-DAPK1:IL23R-Calmodulin complex used for model refinement in Phenix. Additional map: deepEMhancer sharpened map. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Cytokine / Receptor / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprolactin receptor activity / late endosome lumen / interleukin-23 receptor binding / interleukin-12 alpha subunit binding / interleukin-12 complex / interleukin-23 complex / natural killer cell activation involved in immune response / negative regulation of vascular endothelial growth factor signaling pathway / positive regulation of natural killer cell activation / positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target ...prolactin receptor activity / late endosome lumen / interleukin-23 receptor binding / interleukin-12 alpha subunit binding / interleukin-12 complex / interleukin-23 complex / natural killer cell activation involved in immune response / negative regulation of vascular endothelial growth factor signaling pathway / positive regulation of natural killer cell activation / positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target / negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis / positive regulation of lymphocyte proliferation / positive regulation of tissue remodeling / cellular response to hydroperoxide / tissue remodeling / sexual reproduction / regulation of response to tumor cell / positive regulation of autophagic cell death / DAPK1-calmodulin complex / positive regulation of NK T cell activation / positive regulation of T-helper 1 type immune response / positive regulation of smooth muscle cell apoptotic process / positive regulation of mononuclear cell proliferation / interleukin-12 receptor binding / interleukin-12 receptor complex / interleukin-23 receptor complex / T-helper cell differentiation / positive regulation of memory T cell differentiation / interleukin-23-mediated signaling pathway / Interleukin-23 signaling / Caspase activation via Dependence Receptors in the absence of ligand / positive regulation of T-helper 17 type immune response / interleukin-12-mediated signaling pathway / defense response to tumor cell / positive regulation of NK T cell proliferation / negative regulation of interleukin-17 production / Interleukin-12 signaling / positive regulation of osteoclast differentiation / calcium/calmodulin-dependent protein kinase activity / cytokine receptor activity / regulation of NMDA receptor activity / cell surface receptor signaling pathway via STAT / natural killer cell activation / positive regulation of granulocyte macrophage colony-stimulating factor production / response to UV-B / positive regulation of neutrophil chemotaxis / syntaxin-1 binding / CaM pathway / Cam-PDE 1 activation / T-helper 1 type immune response / Sodium/Calcium exchangers / Calmodulin induced events / Reduction of cytosolic Ca++ levels / Activation of Ca-permeable Kainate Receptor / CREB1 phosphorylation through the activation of CaMKII/CaMKK/CaMKIV cascasde / Loss of phosphorylation of MECP2 at T308 / CREB1 phosphorylation through the activation of Adenylate Cyclase / negative regulation of high voltage-gated calcium channel activity / PKA activation / CaMK IV-mediated phosphorylation of CREB / negative regulation of interleukin-10 production / Glycogen breakdown (glycogenolysis) / CLEC7A (Dectin-1) induces NFAT activation / defense response to protozoan / cytokine binding / Activation of RAC1 downstream of NMDARs / negative regulation of ryanodine-sensitive calcium-release channel activity / organelle localization by membrane tethering / mitochondrion-endoplasmic reticulum membrane tethering / autophagosome membrane docking / negative regulation of calcium ion export across plasma membrane / regulation of cardiac muscle cell action potential / presynaptic endocytosis / Interleukin-10 signaling / regulation of cell communication by electrical coupling involved in cardiac conduction / Synthesis of IP3 and IP4 in the cytosol / positive regulation of interleukin-17 production / Phase 0 - rapid depolarisation / calcineurin-mediated signaling / Negative regulation of NMDA receptor-mediated neuronal transmission / Unblocking of NMDA receptors, glutamate binding and activation / RHO GTPases activate PAKs / peptide hormone binding / positive regulation of natural killer cell proliferation / regulation of ryanodine-sensitive calcium-release channel activity / Ion transport by P-type ATPases / positive regulation of activated T cell proliferation / Uptake and function of anthrax toxins / positive regulation of interleukin-10 production / Long-term potentiation / protein phosphatase activator activity / extrinsic apoptotic signaling pathway via death domain receptors / Calcineurin activates NFAT / Regulation of MECP2 expression and activity / negative regulation of protein secretion / DARPP-32 events / Smooth Muscle Contraction / cell surface receptor signaling pathway via JAK-STAT / detection of calcium ion / response to type II interferon Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
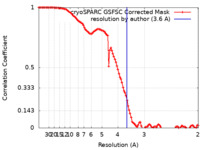
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Bloch Y / Felix J / Savvides SN | |||||||||
| Funding support |  Belgium, 2 items Belgium, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23. Authors: Yehudi Bloch / Jan Felix / Romain Merceron / Mathias Provost / Royan Alipour Symakani / Robin De Backer / Elisabeth Lambert / Ahmad R Mehdipour / Savvas N Savvides /     Abstract: Cell-surface receptor complexes mediated by pro-inflammatory interleukin (IL)-12 and IL-23, both validated therapeutic targets, are incompletely understood due to the lack of structural insights into ...Cell-surface receptor complexes mediated by pro-inflammatory interleukin (IL)-12 and IL-23, both validated therapeutic targets, are incompletely understood due to the lack of structural insights into their complete extracellular assemblies. Furthermore, there is a paucity of structural details describing the IL-12-receptor interaction interfaces, in contrast to IL-23-receptor complexes. Here we report structures of fully assembled mouse IL-12/human IL-23-receptor complexes comprising the complete extracellular segments of the cognate receptors determined by electron cryo-microscopy. The structures reveal key commonalities but also surprisingly diverse features. Most notably, whereas IL-12 and IL-23 both utilize a conspicuously presented aromatic residue on their α-subunit as a hotspot to interact with the N-terminal Ig domain of their high-affinity receptors, only IL-12 juxtaposes receptor domains proximal to the cell membrane. Collectively, our findings will help to complete our understanding of cytokine-mediated assemblies of tall cytokine receptors and will enable a cytokine-specific interrogation of IL-12/IL-23 signaling in physiology and disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16824.map.gz emd_16824.map.gz | 116.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16824-v30.xml emd-16824-v30.xml emd-16824.xml emd-16824.xml | 24.7 KB 24.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16824_fsc.xml emd_16824_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_16824.png emd_16824.png | 72.8 KB | ||
| Masks |  emd_16824_msk_1.map emd_16824_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16824.cif.gz emd-16824.cif.gz | 7.7 KB | ||
| Others |  emd_16824_additional_1.map.gz emd_16824_additional_1.map.gz emd_16824_half_map_1.map.gz emd_16824_half_map_1.map.gz emd_16824_half_map_2.map.gz emd_16824_half_map_2.map.gz | 107.7 MB 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16824 http://ftp.pdbj.org/pub/emdb/structures/EMD-16824 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16824 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16824 | HTTPS FTP |
-Related structure data
| Related structure data |  8oe4MC  8c7mC  8cr5C  8cr6C  8cr8C  8odxC  8odzC  8oe0C  8pb1C  8ppmC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16824.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16824.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map: Unfiltered, non-sharpened map of the human IL23:IL12Rbeta1-DAPK1:IL23R-Calmodulin complex used for model refinement in Phenix. Additional map: deepEMhancer sharpened map. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9815 Å | ||||||||||||||||||||||||||||||||||||
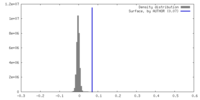
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16824_msk_1.map emd_16824_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
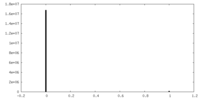
| Projections & Slices |
| ||||||||||||
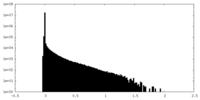
| Density Histograms |
-Additional map: DeepEMhancer sharpened map of the human IL23:IL12Rbeta1-DAPK1:IL23R-Calmodulin complex,...
| File | emd_16824_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer sharpened map of the human IL23:IL12Rbeta1-DAPK1:IL23R-Calmodulin complex, used for model building in Coot and visualization. | ||||||||||||
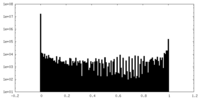
| Projections & Slices |
| ||||||||||||
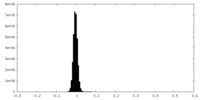
| Density Histograms |
-Half map: Half map 2 of the human IL23:IL12Rbeta1-DAPK1:IL23R-Calmodulin complex....
| File | emd_16824_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of the human IL23:IL12Rbeta1-DAPK1:IL23R-Calmodulin complex. | ||||||||||||
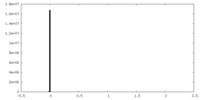
| Projections & Slices |
| ||||||||||||
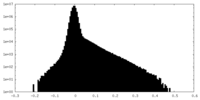
| Density Histograms |
-Half map: Half map 1 of the human IL23:IL12Rbeta1-DAPK1:IL23R-Calmodulin complex....
| File | emd_16824_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of the human IL23:IL12Rbeta1-DAPK1:IL23R-Calmodulin complex. | ||||||||||||
| Projections & Slices |
| ||||||||||||
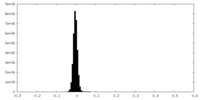
| Density Histograms |
- Sample components
Sample components
-Entire : Human IL-23 in complex with hIL-12Rbeta1-DAPK1 and hIL-23R-Calmodulin.
| Entire | Name: Human IL-23 in complex with hIL-12Rbeta1-DAPK1 and hIL-23R-Calmodulin. |
|---|---|
| Components |
|
-Supramolecule #1: Human IL-23 in complex with hIL-12Rbeta1-DAPK1 and hIL-23R-Calmodulin.
| Supramolecule | Name: Human IL-23 in complex with hIL-12Rbeta1-DAPK1 and hIL-23R-Calmodulin. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 168 KDa |
-Macromolecule #1: Interleukin-12 subunit beta
| Macromolecule | Name: Interleukin-12 subunit beta / type: protein_or_peptide / ID: 1 Details: N-glycosylation mutant: Asparagine 303 (N303) is mutated to Aspartic acid (D303). Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.74093 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: IWELKKDVYV VELDWYPDAP GEMVVLTCDT PEEDGITWTL DQSSEVLGSG KTLTIQVKEF GDAGQYTCHK GGEVLSHSLL LLHKKEDGI WSTDILKDQK EPKNKTFLRC EAKNYSGRFT CWWLTTISTD LTFSVKSSRG SSDPQGVTCG AATLSAERVR G DNKEYEYS ...String: IWELKKDVYV VELDWYPDAP GEMVVLTCDT PEEDGITWTL DQSSEVLGSG KTLTIQVKEF GDAGQYTCHK GGEVLSHSLL LLHKKEDGI WSTDILKDQK EPKNKTFLRC EAKNYSGRFT CWWLTTISTD LTFSVKSSRG SSDPQGVTCG AATLSAERVR G DNKEYEYS VECQEDSACP AAEESLPIEV MVDAVHKLKY ENYTSSFFIR DIIKPDPPKN LQLKPLKNSR QVEVSWEYPD TW STPHSYF SLTFCVQVQG KSKREKKDRV FTDKTSATVI CRKDASISVR AQDRYYSSSW SEWASVPCS UniProtKB: Interleukin-12 subunit beta |
-Macromolecule #2: Interleukin-23 subunit alpha
| Macromolecule | Name: Interleukin-23 subunit alpha / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 20.650117 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RAVPGGSSPA WTQCQQLSQK LCTLAWSAHP LVGHMDLREE GDEETTNDVP HIQCGDGCDP QGLRDNSQFC LQRIHQGLIF YEKLLGSDI FTGEPSLLPD SPVGQLHASL LGLSQLLQPE GHHWETQQIP SLSPSQPWQR LLLRFKILRS LQAFVAVAAR V FAHGAATL SPGDEVDGHH HHHHHHHH UniProtKB: Interleukin-23 subunit alpha |
-Macromolecule #3: Interleukin-23 receptor,Calmodulin-1
| Macromolecule | Name: Interleukin-23 receptor,Calmodulin-1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.991391 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GITNINCSGH IWVEPATIFK MGMNISIYCQ AAIKNCQPRK LHFYKNGIKE RFQITRINKT TARLWYKNFL EPHASMYCTA ECPKHFQET LICGKDISSG YPPDIPDEVT CVIYEYSGNM TCTWNAGKLT YIDTKYVVHV KSLETEEEQQ YLTSSYINIS T DSLQGGKK ...String: GITNINCSGH IWVEPATIFK MGMNISIYCQ AAIKNCQPRK LHFYKNGIKE RFQITRINKT TARLWYKNFL EPHASMYCTA ECPKHFQET LICGKDISSG YPPDIPDEVT CVIYEYSGNM TCTWNAGKLT YIDTKYVVHV KSLETEEEQQ YLTSSYINIS T DSLQGGKK YLVWVQAANA LGMEESKQLQ IHLDDIVIPS AAVISRAETI NATVPKTIIY WDSQTTIEKV SCEMRYKATT NQ TWNVKEF DTNFTYVQQS EFYLEPNIKY VFQVRCQETG KRYWQPWSSL FFHKTPETVP QVTSKAFQHD TWNSGLTVAS IST GHLTSD NRGDGTGGSG GSGGLTEEQI AEFKEAFSLF DKDGDGTITT KELGTVMRSL GQNPTEAELQ DMINEVDADG NGTI DFPEF LTMMARKMKD TDSEEEIREA FRVFDKDGNG YISAAELRHV MTNLGEKLTD EEVDEMIREA DIDGDGQVNY EEFVQ MMTA K UniProtKB: Interleukin-23 receptor, Calmodulin-1 |
-Macromolecule #4: Interleukin-12 receptor subunit beta-1,Death-associated protein k...
| Macromolecule | Name: Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.187316 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: CRTSECCFQD PPYPDADSGS ASGPRDLRCY RISSDRYECS WQYEGPTAGV SHFLRCCLSS GRCCYFAAGS ATRLQFSDQA GVSVLYTVT LWVESWARNQ TEKSPEVTLQ LYNSVKYEPP LGDIKVSKLA GQLRMEWETP DNQVGAEVQF RHRTPSSPWK L GDCGPQDD ...String: CRTSECCFQD PPYPDADSGS ASGPRDLRCY RISSDRYECS WQYEGPTAGV SHFLRCCLSS GRCCYFAAGS ATRLQFSDQA GVSVLYTVT LWVESWARNQ TEKSPEVTLQ LYNSVKYEPP LGDIKVSKLA GQLRMEWETP DNQVGAEVQF RHRTPSSPWK L GDCGPQDD DTESCLCPLE MNVAQEFQLR RRQLGSQGSS WSKWSSPVCV PPENPPQPQV RFSVEQLGQD GRRRLTLKEQ PT QLELPEG CQGLAPGTEV TYRLQLHMLS CPCKAKATRT LHLGKMPYLS GAAYNVAVIS SNQFGPGLNQ TWHIPADTHT EPV ALNISV GTNGTTMYWP ARAQSMTYCI EWQPVGQDGG LATCSLTAPQ DPDPAGMATY SWSRESGAMG QEKCYYITIF ASAH PEKLT LWSTVLSTYH FGGNASAAGT PHHVSVKNHS LDSVSVDWAP SLLSTCPGVL KEYVVRCRDE DSKQVSEHPV QPTET QVTL SGLRAGVAYT VQVRADTAWL RGVWSQPQRF SIEGTGGSGG SGGAARKKWK QSVRLISLCQ RLS UniProtKB: Interleukin-12 receptor subunit beta-1, Death-associated protein kinase 1 |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
Details: HEPES-buffered saline (HBS) with added calcium chloride: 25 mM HEPES, pH 7.4, 150 mM NaCl, 5 mM CaCl | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: LEICA PLUNGER / Details: Leica EM GP2, 4.5 s. blotting time.. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 6660 / Average exposure time: 3.37 sec. / Average electron dose: 61.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 60000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8oe4: |
 Movie
Movie Controller
Controller










































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)