[English] 日本語
 Yorodumi
Yorodumi- EMDB-16404: Subtomogram averaging of a coronavirus spike (ChAdOx1 19E6) in C1... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram averaging of a coronavirus spike (ChAdOx1 19E6) in C1 symmetry | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Coronavirus spike / subtomogram averaging / C1 symmetry / in situ / ChAdOx / VIRAL PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 9.7 Å | |||||||||
 Authors Authors | Ni T / Mendonca L / Zhu Y / Zhang P | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: iScience / Year: 2023 Journal: iScience / Year: 2023Title: ChAdOx1 COVID vaccines express RBD open prefusion SARS-CoV-2 spikes on the cell surface. Authors: Tao Ni / Luiza Mendonça / Yanan Zhu / Andrew Howe / Julika Radecke / Pranav M Shah / Yuewen Sheng / Anna-Sophia Krebs / Helen M E Duyvesteyn / Elizabeth Allen / Teresa Lambe / Cameron ...Authors: Tao Ni / Luiza Mendonça / Yanan Zhu / Andrew Howe / Julika Radecke / Pranav M Shah / Yuewen Sheng / Anna-Sophia Krebs / Helen M E Duyvesteyn / Elizabeth Allen / Teresa Lambe / Cameron Bisset / Alexandra Spencer / Susan Morris / David I Stuart / Sarah Gilbert / Peijun Zhang /  Abstract: Vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been proven to be an effective means of decreasing COVID-19 mortality, hospitalization rates, and transmission. One ...Vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been proven to be an effective means of decreasing COVID-19 mortality, hospitalization rates, and transmission. One of the vaccines deployed worldwide is ChAdOx1 nCoV-19, which uses an adenovirus vector to drive the expression of the original SARS-CoV-2 spike on the surface of transduced cells. Using cryo-electron tomography and subtomogram averaging, we determined the native structures of the vaccine product expressed on cell surfaces . We show that ChAdOx1-vectored vaccines expressing the Beta SARS-CoV-2 variant produce abundant native prefusion spikes predominantly in one-RBD-up conformation. Furthermore, the ChAdOx1-vectored HexaPro-stabilized spike yields higher cell surface expression, enhanced RBD exposure, and reduced shedding of S1 compared to the wild type. We demonstrate structure determination as a powerful means for studying antigen design options in future vaccine development against emerging novel SARS-CoV-2 variants and broadly against other infectious viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16404.map.gz emd_16404.map.gz | 1.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16404-v30.xml emd-16404-v30.xml emd-16404.xml emd-16404.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16404.png emd_16404.png | 96.1 KB | ||
| Filedesc metadata |  emd-16404.cif.gz emd-16404.cif.gz | 4 KB | ||
| Others |  emd_16404_half_map_1.map.gz emd_16404_half_map_1.map.gz emd_16404_half_map_2.map.gz emd_16404_half_map_2.map.gz | 5.3 MB 5.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16404 http://ftp.pdbj.org/pub/emdb/structures/EMD-16404 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16404 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16404 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16404.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16404.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_16404_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
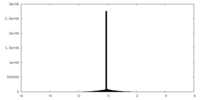
| Density Histograms |
-Half map: #2
| File | emd_16404_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
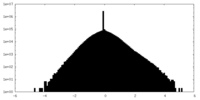
| Density Histograms |
- Sample components
Sample components
-Entire : ChAdOx1 coronavirus spike vaccine carrying hexa-proline mutation
| Entire | Name: ChAdOx1 coronavirus spike vaccine carrying hexa-proline mutation |
|---|---|
| Components |
|
-Supramolecule #1: ChAdOx1 coronavirus spike vaccine carrying hexa-proline mutation
| Supramolecule | Name: ChAdOx1 coronavirus spike vaccine carrying hexa-proline mutation type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 3.17 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 6.5 µm / Nominal defocus min: 2.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 9.7 Å / Resolution method: FSC 0.143 CUT-OFF / Software: (Name: emClarity, RELION) / Number subtomograms used: 28896 |
|---|---|
| Extraction | Number tomograms: 28896 / Number images used: 28896 |
| Final angle assignment | Type: OTHER / Software - Name: emClarity |
 Movie
Movie Controller
Controller


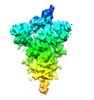




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)