-Search query
-Search result
Showing 1 - 50 of 83 items for (author: kaila & v)

EMDB-17210: 
Respiratory supercomplex (III2-IV2) from Mycobacterium smegmatis
Method: single particle / : Kovalova T, Krol S, Sjostrand D, Riepl D, Gamiz-Hernandez A, Brzezinski P, Kaila V, Hogbom M

EMDB-17211: 
Respiratory supercomplex (III2-IV2) from Mycobacterium smegmatis
Method: single particle / : Kovalova T, Krol S, Sjostrand D, Riepl D, Gamiz-Hernandez A, Brzezinski P, Kaila V, Hogbom M

PDB-8ovc: 
Respiratory supercomplex (III2-IV2) from Mycobacterium smegmatis
Method: single particle / : Kovalova T, Krol S, Sjostrand D, Riepl D, Gamiz-Hernandez A, Brzezinski P, Kaila V, Hogbom M

PDB-8ovd: 
Respiratory supercomplex (III2-IV2) from Mycobacterium smegmatis
Method: single particle / : Kovalova T, Krol S, Sjostrand D, Riepl D, Gamiz-Hernandez A, Brzezinski P, Kaila V, Hogbom M
EMDB-28726: 
Rat 80S ribosome purified from brain RNA granules. Class 1 40S subunit 2_5A resolution
Method: single particle / : Ortega J
EMDB-28727: 
Rat 80S ribosome purified from brain RNA granules. Class 1 60S subunit 2_5A resolution.
Method: single particle / : Ortega J
EMDB-26517: 
Rat 80S ribosome purified from brain RNA granules. Class 2 40S subunit.
Method: single particle / : Ortega J
EMDB-26518: 
Rat 80S ribosome purified from brain RNA granules. Class 2 60S subunit.
Method: single particle / : Ortega J
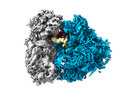
EMDB-29538: 
Rat 80S ribosome purified from brain RNA granules. Class 1 80S consensus
Method: single particle / : Ortega J
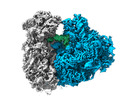
EMDB-29539: 
Rat 80S ribosome purified from brain RNA granules. Class 2 80S consensus
Method: single particle / : Ortega J

EMDB-13818: 
Electron bifurcating hydrogenase - HydABC from A. woodii
Method: single particle / : Katsyv A, Kumar A, Saura P, Poeverlein MC, Freibert SA, Stripp S, Jain S, Gamiz-Hernandez AP, Kaila VRI, Mueller V, Schuller JM

EMDB-15166: 
Cryo-EM structure of the electron bifurcating Fe-Fe hydrogenase HydABC complex from Acetobacterium woodii in the reduced state
Method: single particle / : Kumar A, Saura P, Gamiz-Hernandez AP, Kaila VRI, Mueller V, Schuller JM

PDB-7q4v: 
Electron bifurcating hydrogenase - HydABC from A. woodii
Method: single particle / : Katsyv A, Kumar A, Saura P, Poeverlein MC, Freibert SA, Stripp S, Jain S, Gamiz-Hernandez AP, Kaila VRI, Mueller V, Schuller JM

PDB-8a5e: 
Cryo-EM structure of the electron bifurcating Fe-Fe hydrogenase HydABC complex from Acetobacterium woodii in the reduced state
Method: single particle / : Kumar A, Saura P, Gamiz-Hernandez AP, Kaila VRI, Mueller V, Schuller JM

EMDB-13819: 
CryoEM structure of electron bifurcating Fe-Fe hydrogenase HydABC complex A. woodii in the oxidised state
Method: single particle / : Kumar A, Saura P, Poeverlein MC, Gamiz-Hernandez AP, Kaila VRI, Mueller V, Schuller JM

EMDB-15212: 
Cryo-EM structure of the electron bifurcating Fe-Fe hydrogenase HydABC complex from Thermoanaerobacter kivui in the reduced state
Method: single particle / : Kumar A, Saura P, Gamiz-Hernandez AP, Kaila VRI, Mueller V, Schuller JM

EMDB-16011: 
Cryo-EM structure of the electron bifurcating Fe-Fe hydrogenase HydABC complex from Thermoanaerobacter kivui in the oxidised state
Method: single particle / : Kumar A, Schuller JM

PDB-7q4w: 
CryoEM structure of electron bifurcating Fe-Fe hydrogenase HydABC complex A. woodii in the oxidised state
Method: single particle / : Kumar A, Saura P, Poeverlein MC, Gamiz-Hernandez AP, Kaila VRI, Mueller V, Schuller JM

PDB-8a6t: 
Cryo-EM structure of the electron bifurcating Fe-Fe hydrogenase HydABC complex from Thermoanaerobacter kivui in the reduced state
Method: single particle / : Kumar A, Saura P, Gamiz-Hernandez AP, Kaila VRI, Mueller V, Schuller JM

PDB-8bew: 
Cryo-EM structure of the electron bifurcating Fe-Fe hydrogenase HydABC complex from Thermoanaerobacter kivui in the oxidised state
Method: single particle / : Kumar A, Schuller JM

EMDB-27196: 
Mammalian CIV with GDN bound
Method: single particle / : Di Trani J, Rubinstein J

PDB-8d4t: 
Mammalian CIV with GDN bound
Method: single particle / : Di Trani J, Rubinstein J

EMDB-23831: 
Human CTPS1 bound to UTP, AMPPNP, and glutamine
Method: single particle / : Lynch EM, Dimattia MA

EMDB-23832: 
Human CTPS1 bound to CTP
Method: single particle / : Lynch EM, Kollman JM

EMDB-23833: 
Human CTPS2 bound to CTP
Method: single particle / : Lynch EM, Kollman JM

EMDB-23848: 
Human CTPS1 bound to inhibitor R80
Method: single particle / : Lynch EM, Dimattia MA, Kollman JM

EMDB-23850: 
Human CTPS1 bound to inhibitor T35
Method: single particle / : Lynch EM, Dimattia MA, Kollman JM

EMDB-23851: 
Human CTPS2 bound to inhibitor R80
Method: single particle / : Lynch EM, Dimattia MA

EMDB-23852: 
Human CTPS2 bound to inhibitor T35
Method: single particle / : Lynch EM, Dimattia MA

EMDB-23859: 
Mouse CTPS1 bound to inhibitor R80
Method: single particle / : Lynch EM, Dimattia MA, Kollman JM

EMDB-23865: 
Mouse CTPS2 bound to inhibitor R80
Method: single particle / : Lynch EM, Dimattia MA

EMDB-23866: 
Mouse CTPS2-I250T bound to inhibitor R80
Method: single particle / : Lynch EM, Dimattia MA

PDB-7mgz: 
Human CTPS1 bound to UTP, AMPPNP, and glutamine
Method: single particle / : Lynch EM, Dimattia MA, Kollman JM

PDB-7mh0: 
Human CTPS1 bound to CTP
Method: single particle / : Lynch EM, Kollman JM

PDB-7mh1: 
Human CTPS2 bound to CTP
Method: single particle / : Lynch EM, Kollman JM

PDB-7mif: 
Human CTPS1 bound to inhibitor R80
Method: single particle / : Lynch EM, Dimattia MA, Kollman JM

PDB-7mig: 
Human CTPS1 bound to inhibitor T35
Method: single particle / : Lynch EM, Dimattia MA, Kollman JM

PDB-7mih: 
Human CTPS2 bound to inhibitor R80
Method: single particle / : Lynch EM, Dimattia MA, Kollman JM

PDB-7mii: 
Human CTPS2 bound to inhibitor T35
Method: single particle / : Lynch EM, Dimattia MA, Kollman JM

PDB-7mip: 
Mouse CTPS1 bound to inhibitor R80
Method: single particle / : Lynch EM, Dimattia MA, Kollman JM

PDB-7miu: 
Mouse CTPS2 bound to inhibitor R80
Method: single particle / : Lynch EM, Dimattia MA, Kollman JM

PDB-7miv: 
Mouse CTPS2-I250T bound to inhibitor R80
Method: single particle / : Lynch EM, Dimattia MA, Kollman JM

EMDB-12885: 
Structure of the apo-state of the bacteriophage PhiKZ non-virion RNA polymerase - class including clamp
Method: single particle / : de Martin Garrido N, Lai Wan Loong YTE

EMDB-12886: 
Structure of the apo-state of the bacteriophage PhiKZ non-virion RNA polymerase
Method: single particle / : de Martin Garrido N, Lai Wan Loong YTE

PDB-7ogp: 
Structure of the apo-state of the bacteriophage PhiKZ non-virion RNA polymerase - class including clamp
Method: single particle / : de Martin Garrido N, Lai Wan Loong YTE, Yakunina M, Aylett CHS

PDB-7ogr: 
Structure of the apo-state of the bacteriophage PhiKZ non-virion RNA polymerase
Method: single particle / : de Martin Garrido N, Lai Wan Loong YTE, Yakunina M, Aylett CHS
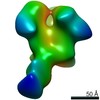
EMDB-22496: 
Base-directed response of rabbit #1 week 6 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
Method: single particle / : Sewell LM, Ozorowski G, Ward AB

EMDB-22498: 
N611-site directed response of rabbit #1 week 6 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
Method: single particle / : Sewell LM, Ozorowski G, Ward AB

EMDB-22499: 
Base-directed response of rabbit #1 week 12 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
Method: single particle / : Sewell LM, Ozorowski G, Ward AB
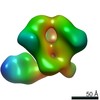
EMDB-22500: 
N611-site directed response of rabbit #1 week 12 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
Method: single particle / : Sewell LM, Ozorowski G, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
